Difference between revisions of "Journal:Launching genomics into the cloud: Deployment of Mercury, a next generation sequence analysis pipeline"
Shawndouglas (talk | contribs) m (Fixed DOI) |
Shawndouglas (talk | contribs) (Added content. Saving and adding more.) |
||
| Line 35: | Line 35: | ||
'''Keywords:''' NGS data, Variant calling, Annotation, Clinical sequencing, Cloud computing | '''Keywords:''' NGS data, Variant calling, Annotation, Clinical sequencing, Cloud computing | ||
==Background== | |||
Whole exome capture sequencing (WES) and whole genome sequencing (WGS) using next generation sequencing (NGS) technologies<ref name="MetzkerSeq10">{{cite journal |title=Sequencing technologies — The next generation |journal=Nature Reviews Genetics |author=Metzker, Michael L. |volume=11 |issue=1 |pages=31–46 |year=2010 |doi=10.1038/nrg2626 |pmid=19997069}}</ref> have emerged as compelling paradigms for routine clinical diagnosis, genetic risk prediction, and patient management.<ref name="BainbridgeWhole11">{{cite journal |title=Whole-genome sequencing for optimized patient management |journal=Science Translational Medicine |author=Bainbridge, Matthew N.; Wiszniewski, Wojciech; Murdock, David R. et al. |volume=3 |issue=87 |pages=87re3 |year=2011 |doi=10.1126/scitranslmed.3002243 |pmid=21677200 |pmc=PMC3314311}}</ref> Large numbers of laboratories and hospitals routinely generate terabytes of NGS data, shifting the bottleneck in clinical genetics from DNA sequence production to DNA sequence analysis. Such analysis is most prevalent in three common settings: first, in a clinical diagnostics laboratory (e.g. Baylor’s Whole Genome Laboratory http://www.bcm.edu/geneticlabs/) testing single patients or families with presumed heritable disease; second, in a cancer-analysis setting where the unit of interest is either a normal-tumor tissue pair or normal-primary tumor-recurrence trio<ref name="CancerInt11">{{cite journal |title=Integrated genomic analyses of ovarian carcinoma |journal=Nature |author=Cancer Genome Atlas Research Network |volume=474 |issue=7353 |pages=609–615 |year=2011 |doi=10.1038/nature10166 |pmid=21720365 |pmc=PMC3163504}}</ref>; and third, in biomedical research studies sequencing a sample of well-phenotyped individuals. In each case, the input is a DNA sample of appropriate quality having a unique identification number, appropriate informed consent, and relevant clinical and phenotypic information. | |||
As these new samples are sequenced, the resulting data is most effectively examined in the context of petabases of existing DNA sequence and the associated meta-data. Such large-scale comparative [[genomics]] requires new sequence data to be robustly characterized, consistently reproducible, and easily shared among large collaborations in a secure manner. And while data-management and [[information]] technologies have adapted to the processing and storage requirements of emerging sequencing technologies (e.g., the CRAM specification<ref name="WheelerTheCom08">{{cite journal |title=The complete genome of an individual by massively parallel DNA sequencing |journal=Nature |author=Wheeler, David A.; Srinivasan, Maithreyan; Egholm, Michael et al. |volume=452 |issue=7189 |pages=872–876 |year=2008 |doi=10.1038/nature06884 |pmid=18421352}}</ref>), it is less certain that appropriate informative software interfaces will be made available to the genomics and clinical genetics communities. One element bridging the technology gap between the sequencing instrument and the scientist or clinician is a validated data processing pipeline that takes raw sequencing reads and produces an annotated personal genome ready for further analysis and clinical interpretation. | |||
To address this need, we have designed and implemented Mercury, an automated approach that integrates multiple sequence analysis components across many computational steps, from obtaining patient samples to providing a fully annotated list of variant sites for clinical applications. Mercury fully integrates new software with existing routines (e.g., Altas2<ref name="ChallisAnInt12">{{cite journal |title=An integrative variant analysis suite for whole exome next-generation sequencing data |journal=BMC Bioinformatics |author=Challis, D.; Yu, J.; Evani, U.S. et al. |volume=13 |pages=8 |year=2012 |doi=10.1186/1471-2105-13-8 |pmid=22239737 |pmc=PMC3292476}}</ref>) and provides the flexibility necessary to respond to changing sequencing technologies and the rapidly increasing volume of relevant data. Mercury has been implemented on both local infrastructure and in a cloud computing platform provided by DNAnexus using Amazon Web Services (AWS). While there are other NGS analysis pipelines, some of which have even been implemented in the cloud<ref name="ODriscollBig13">{{cite journal |title='Big data', Hadoop and cloud computing in genomics |journal=Journal of Biomedical Informatics |author=O’Driscoll, A.; Daugelaite, J.; Sleator, R.D. |volume=46 |issue=5 |pages=774-81 |year=2013 |doi=10.1016/j.jbi.2013.07.001 |pmid=23872175}}</ref>, the combination of Mercury and DNAnexus together provide for the first time a fully integrated genomic analysis resource that can serve the full spectrum of users. | |||
==Results and discussion== | |||
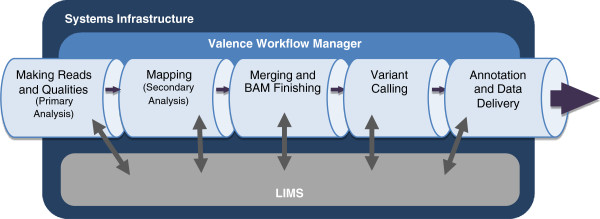
Figure 1 provides an overview of the Mercury data processing pipeline. Source information includes sample and project management data and the characteristics of library preparation and sequencing. This information enters the pipeline either directly from the user or from a laboratory information management system (LIMS). The first step, generating sequencing reads, is based on the vendor’s primary analysis software, which generates sequence reads and base-call confidence values (qualities). The second step maps the reads and qualities to the reference genome using a standard mapping tool, such as BWA<ref name="LiFastShort09">{{cite journal |title=Fast and accurate short read alignment with Burrows–Wheeler transform |journal=Bioinformatics |author=Li, H.; Durbin, R. |volume=25 |issue=14 |pages=1754–60 |year=2009 |doi=10.1093/bioinformatics/btp324 |pmid=19451168 |pmc=PMC2705234}}</ref><ref name="LiFastLong09">{{cite journal |title=Fast and accurate long-read alignment with Burrows–Wheeler transform |journal=Bioinformatics |author=Li, H.; Durbin, R. |volume=26 |issue=5 |pages=589-95 |year=2010 |doi=10.1093/bioinformatics/btp698 |pmid=20080505 |pmc=PMC2828108}}</ref>, producing a BAM<ref name="LiTheSeq09">{{cite journal |title=The Sequence Alignment/Map format and SAMtools |journal=Bioinformatics |author=Li, H.; Handsaker, B.; Wysoker, A. et al. |volume=25 |issue=16 |pages=2078-9 |year=2009 |doi=10.1093/bioinformatics/btp352 |pmid=19505943 |pmc=PMC2723002}}</ref> (binary alignment/map) file. The third step produces a “finished” BAM that includes sorting, duplicate marking, indel realignment, base quality recalibration, and indexing (using a combination of tools including SAMtools<ref name="LiTheSeq09" />, Picard (http://picard.sourceforge.net), and GATK<ref name="DePristoAFrame11">{{cite journal |title=A framework for variation discovery and genotyping using next-generation DNA sequencing data |journal=Nature Genetics |author=DePristo, M.A.; Banks, E.; Poplin, R. et al. |volume=43 |issue=5 |pages=491-8 |year=2011 |doi=10.1038/ng.806 |pmid=21478889 |pmc=PMC3083463}}</ref>). The fourth step in Mercury uses the Atlas2 suite<ref name="ChallisAnInt12" /><ref name="ShenSNP10">{{cite journal |title=A SNP discovery method to assess variant allele probability from next-generation resequencing data |journal=Genome Research |author=Shen, Y.; Wan, Z.; Coarfa, C. et al. |volume=20 |issue=2 |pages=273-80 |year=2010 |doi=10.1101/gr.096388.109 |pmid=20019143 |pmc=PMC2813483}}</ref> (Atlas-SNP and Atlas-indel) to call variants and produce a variant file (VCF). The fifth step adds biological and functional annotation and formats the variant lists for delivery. Each step is described in detail in the Methods section, as is the flow of information between steps. | |||
[[File:Fig1 Reid BMCInformatics2014 15.jpg|600px]] | |||
{{clear}} | |||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| border="0" cellpadding="5" cellspacing="0" width="600px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 1. Mercury Data Flow.''' 1) Sequencing Instrument raw data is passed to vendor primary analysis software to generate sequence reads and base call confidence values (qualities). 2) Reads and qualities are passed to a mapping tool (BWA) for comparison to a reference genome to determine the placement of reads on the reference (producing a BAM file). 3) Individual sequence event BAMs are merged to make a single sample-level BAM file that then is processed in preparation for variant calling. 4) Atlas-SNP and Atlas-indel are used to identify variants and produce variant files (VCF). 5) Annotation adds biological and functional information to the variant lists and formats them for delivery.</blockquote> | |||
|- | |||
|} | |||
|} | |||
Mercury has been optimized for the Illumina HiSeq (Illumina, Inc.; San Diego, CA) platform, but the generalized workflow framework is adaptable to other settings. The entire pipeline has been implemented both locally and in a cloud computing environment. All relevant code and documentation are freely available online (https://www.hgsc.bcm.edu/content/mercury) and the scalable cloud solution is available within the DNAnexus library (http://www.dnanexus.com/). Sensible default parameters have already been determined so that researchers and clinicians can reliably analyze their data with Mercury without needing to configure any of the constituent programs or obtaining access to large computational resources, and they can do so in a manner compliant with multiple regulatory frameworks. | |||
===Local workflow management=== | |||
Implementing a robust analysis framework that incorporates a heterogeneous collection of software tools presents many challenges. Running disparate software modules with varying inputs and outputs that depend on each other’s results requires appropriate error checking and centralized output logging. We therefore developed a simple yet extensible workflow management framework, Valence (http://sourceforge.net/projects/valence/), that manages the various steps and dependencies within Mercury and handles internal and external pipeline communication. This formal approach to workflow management helps maximize computational resource utilization and seamlessly guides the data from the sequencing instrument to an annotated variant file ready for clinical interpretation and downstream analysis. | |||
Valence parses and executes an analysis protocol described in XML format with each step treated as either an action or a procedure. An action is defined as a direct call to the system to submit a program or script to the job scheduler for execution; a procedure is defined a collection of actions, which is itself a workflow. This design allows the user to easily add, remove, and modify the steps of any analysis protocol. A protocol description for a specific step must include the required cluster resources, any dependencies on other steps, and a command element that describes how to execute the program or script. To ensure that the XML wrappers are applicable to any run, the command is treated as a string template that allows XML variables to be substituted into the command prior to execution. Thus, a single XML wrapper describing how to run a program can be applied to different inputs. Valence can be deployed on any cluster with a job scheduler (e.g., PBS, LSF, SGE), implementing a database to track both the job (the collection of all the steps in a protocol to be executed) and the status (“Not Started,” “Running,” “Finished,” “Failed”) of any action associated with the job. | |||
Mercury users can easily incorporate new analysis tools into an existing pipeline. For example, we recently expanded the scope of our pipeline to include Tiresias (https://sourceforge.net/projects/tiresias/), a structural variant caller focused on mobile elements, and ExCID (https://github.com/cbuhay/ExCID), an exome coverage analysis tool designed to provide clinical reports on under-covered regions. To incorporate Tiresias and ExCID into the Mercury pipeline, we needed only to specify the compute requirements and add the appropriate command to the existing XML workflow definition; Valence then automatically handles all data passing, logging, and error reporting. | |||
===Cloud workflow management=== | |||
Mercury has been instantiated in the cloud via the DNAnexus platform (utilizing AWS’s EC2 and S3). DNAnexus provides a layer of abstraction that simplifies development, execution, monitoring, and collaboration on a cloud infrastructure. The constituent steps of the Mercury pipeline take the form of discrete “applets,” which are then linked to form a workflow within the DNAnexus platform infrastructure. Using the workflow construction GUI, one can add applets (representing each step) to the workflow and create a dependency graph by linking the inputs and outputs of subsequent applets. Inputs are provided to an instance of the workflow, and the entire workflow is run within the cloud infrastructure. The various steps within the workflow are then executed based on the dependency graph. As with Valence, individual applets can be configured to run with a specific set of computational resource requirements such as memory, local disk space, and number of cores and processors. We are currently working to merge the local and cloud infrastructure elements by integrating the upload agent into Valence, allowing Valence to trigger a DNAnexus workflow once all the data is successfully uploaded. Such coordination would serve to transparently support analysis bursts. | |||
The Mercury pipeline within DNAnexus comprises code that uses the DNAnexus command-line interface to instantiate the pipeline in the cloud. The Mercury code for DNAnexus is executed on a local terminal. For example, one may provide a list of sample FASTQ files and sample meta-data locations to Mercury, at which point Mercury uploads the data and instantiates the predefined workflow within DNAnexus. On average, on a 100 Mbps connection, we were able to upload at a rate of ~14 MB/sec. We were able to parallelize this uploading process, yielding an effective upload rate of ~90 MB/sec. The size of a typical FASTQ file from WES with 150X coverage has a compressed (bzip2) file size of approximately 3 GB. Uploading such a file from a local server took less than five minutes. After sample data are uploaded to the DNAnexus environment, the workflow is instantiated in the cloud. | |||
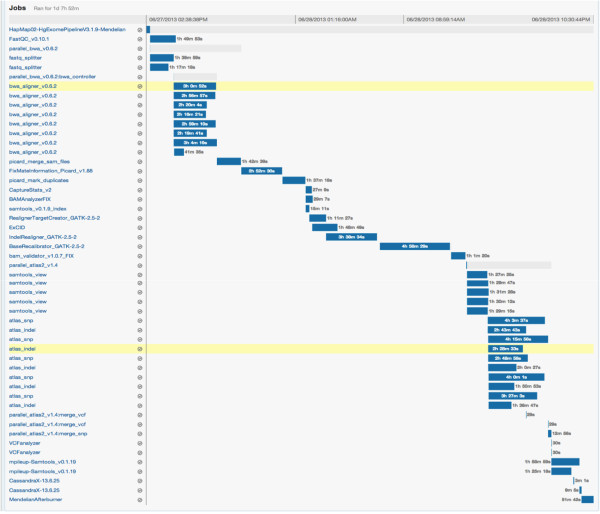
Progress can be monitored online using the DNAnexus GUI (Figure 2) or locally through the Mercury monitoring code. To achieve full automation, the monitoring code can be made a part of a local process to poll for analysis status at regular intervals and start analysis of new sequences automatically upon completion of sequencing. When the Mercury monitoring code detects successful completion of analysis an email notification is sent out. The results can either be downloaded to the local server or the user can view various tracks and data with a native genome browser. | |||
[[File:Fig2 Reid BMCInformatics2014 15.jpg|600px]] | |||
{{clear}} | |||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| border="0" cellpadding="5" cellspacing="0" width="600px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 2. Workflow monitoring in DNAnexus.''' The GUI for applet monitoring displays the progress as a Gantt chart. The left panel lists the various steps including the parallelization steps with each row corresponding to a compute instance. A particular step can be clicked to determine the exact inputs and output or logs of execution for that step. Here we show a snapshot of the webpage displaying the progress of execution for the NA12878 exome analysis. </blockquote> | |||
|- | |||
|} | |||
|} | |||
==References== | ==References== | ||
{{Reflist|colwidth=30em}} | {{Reflist|colwidth=30em}} | ||
Revision as of 18:38, 18 December 2015
| Full article title | Launching genomics into the cloud: Deployment of Mercury, a next generation sequence analysis pipeline |
|---|---|
| Journal | BMC Bioinformatics |
| Author(s) |
Reid, Jeffrey G.; Carroll, Andrew; Veeraraghavan, Narayanan; Dahdouli, Mahmoud; Sundquist, Andreas; English, Adam; Bainbridge, Matthew; White, Simon; Salerno, William; Buhay, Christian; Yu, Fuli; Muzny, Donna; Daly, Richard; Duyk, Geoff; Gibbs, Richard A. Boerwinkle, Eric |
| Author affiliation(s) | Baylor College of Medicine; DNAnexus |
| Primary contact | Email: jgreid@bcm.edu |
| Year published | 2014 |
| Volume and issue | 15 |
| Page(s) | 30 |
| DOI | 10.1186/1471-2105-15-30 |
| ISSN | 1471-2105 |
| Distribution license | Creative Commons Attribution 2.0 Generic |
| Website | http://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-15-30 |
| Download | http://bmcbioinformatics.biomedcentral.com/track/pdf/10.1186/1471-2105-15-30 (PDF) |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
Background: Massively parallel DNA sequencing generates staggering amounts of data. Decreasing cost, increasing throughput, and improved annotation have expanded the diversity of genomics applications in research and clinical practice. This expanding scale creates analytical challenges: accommodating peak compute demand, coordinating secure access for multiple analysts, and sharing validated tools and results.
Results: To address these challenges, we have developed the Mercury analysis pipeline and deployed it in local hardware and the Amazon Web Services cloud via the DNAnexus platform. Mercury is an automated, flexible, and extensible analysis workflow that provides accurate and reproducible genomic results at scales ranging from individuals to large cohorts.
Conclusions: By taking advantage of cloud computing and with Mercury implemented on the DNAnexus platform, we have demonstrated a powerful combination of a robust and fully validated software pipeline and a scalable computational resource that, to date, we have applied to more than 10,000 whole genome and whole exome samples.
Keywords: NGS data, Variant calling, Annotation, Clinical sequencing, Cloud computing
Background
Whole exome capture sequencing (WES) and whole genome sequencing (WGS) using next generation sequencing (NGS) technologies[1] have emerged as compelling paradigms for routine clinical diagnosis, genetic risk prediction, and patient management.[2] Large numbers of laboratories and hospitals routinely generate terabytes of NGS data, shifting the bottleneck in clinical genetics from DNA sequence production to DNA sequence analysis. Such analysis is most prevalent in three common settings: first, in a clinical diagnostics laboratory (e.g. Baylor’s Whole Genome Laboratory http://www.bcm.edu/geneticlabs/) testing single patients or families with presumed heritable disease; second, in a cancer-analysis setting where the unit of interest is either a normal-tumor tissue pair or normal-primary tumor-recurrence trio[3]; and third, in biomedical research studies sequencing a sample of well-phenotyped individuals. In each case, the input is a DNA sample of appropriate quality having a unique identification number, appropriate informed consent, and relevant clinical and phenotypic information.
As these new samples are sequenced, the resulting data is most effectively examined in the context of petabases of existing DNA sequence and the associated meta-data. Such large-scale comparative genomics requires new sequence data to be robustly characterized, consistently reproducible, and easily shared among large collaborations in a secure manner. And while data-management and information technologies have adapted to the processing and storage requirements of emerging sequencing technologies (e.g., the CRAM specification[4]), it is less certain that appropriate informative software interfaces will be made available to the genomics and clinical genetics communities. One element bridging the technology gap between the sequencing instrument and the scientist or clinician is a validated data processing pipeline that takes raw sequencing reads and produces an annotated personal genome ready for further analysis and clinical interpretation.
To address this need, we have designed and implemented Mercury, an automated approach that integrates multiple sequence analysis components across many computational steps, from obtaining patient samples to providing a fully annotated list of variant sites for clinical applications. Mercury fully integrates new software with existing routines (e.g., Altas2[5]) and provides the flexibility necessary to respond to changing sequencing technologies and the rapidly increasing volume of relevant data. Mercury has been implemented on both local infrastructure and in a cloud computing platform provided by DNAnexus using Amazon Web Services (AWS). While there are other NGS analysis pipelines, some of which have even been implemented in the cloud[6], the combination of Mercury and DNAnexus together provide for the first time a fully integrated genomic analysis resource that can serve the full spectrum of users.
Results and discussion
Figure 1 provides an overview of the Mercury data processing pipeline. Source information includes sample and project management data and the characteristics of library preparation and sequencing. This information enters the pipeline either directly from the user or from a laboratory information management system (LIMS). The first step, generating sequencing reads, is based on the vendor’s primary analysis software, which generates sequence reads and base-call confidence values (qualities). The second step maps the reads and qualities to the reference genome using a standard mapping tool, such as BWA[7][8], producing a BAM[9] (binary alignment/map) file. The third step produces a “finished” BAM that includes sorting, duplicate marking, indel realignment, base quality recalibration, and indexing (using a combination of tools including SAMtools[9], Picard (http://picard.sourceforge.net), and GATK[10]). The fourth step in Mercury uses the Atlas2 suite[5][11] (Atlas-SNP and Atlas-indel) to call variants and produce a variant file (VCF). The fifth step adds biological and functional annotation and formats the variant lists for delivery. Each step is described in detail in the Methods section, as is the flow of information between steps.
|
Mercury has been optimized for the Illumina HiSeq (Illumina, Inc.; San Diego, CA) platform, but the generalized workflow framework is adaptable to other settings. The entire pipeline has been implemented both locally and in a cloud computing environment. All relevant code and documentation are freely available online (https://www.hgsc.bcm.edu/content/mercury) and the scalable cloud solution is available within the DNAnexus library (http://www.dnanexus.com/). Sensible default parameters have already been determined so that researchers and clinicians can reliably analyze their data with Mercury without needing to configure any of the constituent programs or obtaining access to large computational resources, and they can do so in a manner compliant with multiple regulatory frameworks.
Local workflow management
Implementing a robust analysis framework that incorporates a heterogeneous collection of software tools presents many challenges. Running disparate software modules with varying inputs and outputs that depend on each other’s results requires appropriate error checking and centralized output logging. We therefore developed a simple yet extensible workflow management framework, Valence (http://sourceforge.net/projects/valence/), that manages the various steps and dependencies within Mercury and handles internal and external pipeline communication. This formal approach to workflow management helps maximize computational resource utilization and seamlessly guides the data from the sequencing instrument to an annotated variant file ready for clinical interpretation and downstream analysis.
Valence parses and executes an analysis protocol described in XML format with each step treated as either an action or a procedure. An action is defined as a direct call to the system to submit a program or script to the job scheduler for execution; a procedure is defined a collection of actions, which is itself a workflow. This design allows the user to easily add, remove, and modify the steps of any analysis protocol. A protocol description for a specific step must include the required cluster resources, any dependencies on other steps, and a command element that describes how to execute the program or script. To ensure that the XML wrappers are applicable to any run, the command is treated as a string template that allows XML variables to be substituted into the command prior to execution. Thus, a single XML wrapper describing how to run a program can be applied to different inputs. Valence can be deployed on any cluster with a job scheduler (e.g., PBS, LSF, SGE), implementing a database to track both the job (the collection of all the steps in a protocol to be executed) and the status (“Not Started,” “Running,” “Finished,” “Failed”) of any action associated with the job.
Mercury users can easily incorporate new analysis tools into an existing pipeline. For example, we recently expanded the scope of our pipeline to include Tiresias (https://sourceforge.net/projects/tiresias/), a structural variant caller focused on mobile elements, and ExCID (https://github.com/cbuhay/ExCID), an exome coverage analysis tool designed to provide clinical reports on under-covered regions. To incorporate Tiresias and ExCID into the Mercury pipeline, we needed only to specify the compute requirements and add the appropriate command to the existing XML workflow definition; Valence then automatically handles all data passing, logging, and error reporting.
Cloud workflow management
Mercury has been instantiated in the cloud via the DNAnexus platform (utilizing AWS’s EC2 and S3). DNAnexus provides a layer of abstraction that simplifies development, execution, monitoring, and collaboration on a cloud infrastructure. The constituent steps of the Mercury pipeline take the form of discrete “applets,” which are then linked to form a workflow within the DNAnexus platform infrastructure. Using the workflow construction GUI, one can add applets (representing each step) to the workflow and create a dependency graph by linking the inputs and outputs of subsequent applets. Inputs are provided to an instance of the workflow, and the entire workflow is run within the cloud infrastructure. The various steps within the workflow are then executed based on the dependency graph. As with Valence, individual applets can be configured to run with a specific set of computational resource requirements such as memory, local disk space, and number of cores and processors. We are currently working to merge the local and cloud infrastructure elements by integrating the upload agent into Valence, allowing Valence to trigger a DNAnexus workflow once all the data is successfully uploaded. Such coordination would serve to transparently support analysis bursts.
The Mercury pipeline within DNAnexus comprises code that uses the DNAnexus command-line interface to instantiate the pipeline in the cloud. The Mercury code for DNAnexus is executed on a local terminal. For example, one may provide a list of sample FASTQ files and sample meta-data locations to Mercury, at which point Mercury uploads the data and instantiates the predefined workflow within DNAnexus. On average, on a 100 Mbps connection, we were able to upload at a rate of ~14 MB/sec. We were able to parallelize this uploading process, yielding an effective upload rate of ~90 MB/sec. The size of a typical FASTQ file from WES with 150X coverage has a compressed (bzip2) file size of approximately 3 GB. Uploading such a file from a local server took less than five minutes. After sample data are uploaded to the DNAnexus environment, the workflow is instantiated in the cloud. Progress can be monitored online using the DNAnexus GUI (Figure 2) or locally through the Mercury monitoring code. To achieve full automation, the monitoring code can be made a part of a local process to poll for analysis status at regular intervals and start analysis of new sequences automatically upon completion of sequencing. When the Mercury monitoring code detects successful completion of analysis an email notification is sent out. The results can either be downloaded to the local server or the user can view various tracks and data with a native genome browser.
|
References
- ↑ Metzker, Michael L. (2010). "Sequencing technologies — The next generation". Nature Reviews Genetics 11 (1): 31–46. doi:10.1038/nrg2626. PMID 19997069.
- ↑ Bainbridge, Matthew N.; Wiszniewski, Wojciech; Murdock, David R. et al. (2011). "Whole-genome sequencing for optimized patient management". Science Translational Medicine 3 (87): 87re3. doi:10.1126/scitranslmed.3002243. PMC PMC3314311. PMID 21677200. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3314311.
- ↑ Cancer Genome Atlas Research Network (2011). "Integrated genomic analyses of ovarian carcinoma". Nature 474 (7353): 609–615. doi:10.1038/nature10166. PMC PMC3163504. PMID 21720365. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3163504.
- ↑ Wheeler, David A.; Srinivasan, Maithreyan; Egholm, Michael et al. (2008). "The complete genome of an individual by massively parallel DNA sequencing". Nature 452 (7189): 872–876. doi:10.1038/nature06884. PMID 18421352.
- ↑ 5.0 5.1 Challis, D.; Yu, J.; Evani, U.S. et al. (2012). "An integrative variant analysis suite for whole exome next-generation sequencing data". BMC Bioinformatics 13: 8. doi:10.1186/1471-2105-13-8. PMC PMC3292476. PMID 22239737. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3292476.
- ↑ O’Driscoll, A.; Daugelaite, J.; Sleator, R.D. (2013). "'Big data', Hadoop and cloud computing in genomics". Journal of Biomedical Informatics 46 (5): 774-81. doi:10.1016/j.jbi.2013.07.001. PMID 23872175.
- ↑ Li, H.; Durbin, R. (2009). "Fast and accurate short read alignment with Burrows–Wheeler transform". Bioinformatics 25 (14): 1754–60. doi:10.1093/bioinformatics/btp324. PMC PMC2705234. PMID 19451168. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2705234.
- ↑ Li, H.; Durbin, R. (2010). "Fast and accurate long-read alignment with Burrows–Wheeler transform". Bioinformatics 26 (5): 589-95. doi:10.1093/bioinformatics/btp698. PMC PMC2828108. PMID 20080505. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2828108.
- ↑ 9.0 9.1 Li, H.; Handsaker, B.; Wysoker, A. et al. (2009). "The Sequence Alignment/Map format and SAMtools". Bioinformatics 25 (16): 2078-9. doi:10.1093/bioinformatics/btp352. PMC PMC2723002. PMID 19505943. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2723002.
- ↑ DePristo, M.A.; Banks, E.; Poplin, R. et al. (2011). "A framework for variation discovery and genotyping using next-generation DNA sequencing data". Nature Genetics 43 (5): 491-8. doi:10.1038/ng.806. PMC PMC3083463. PMID 21478889. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3083463.
- ↑ Shen, Y.; Wan, Z.; Coarfa, C. et al. (2010). "A SNP discovery method to assess variant allele probability from next-generation resequencing data". Genome Research 20 (2): 273-80. doi:10.1101/gr.096388.109. PMC PMC2813483. PMID 20019143. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2813483.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added.