Difference between revisions of "Journal:Method-centered digital communities on protocols.io for fast-paced scientific innovation"
Shawndouglas (talk | contribs) (Created stub. Saving and adding more.) |
Shawndouglas (talk | contribs) (Saving and adding more.) |
||
| Line 43: | Line 43: | ||
===Adding protocols in protocols.io=== | ===Adding protocols in protocols.io=== | ||
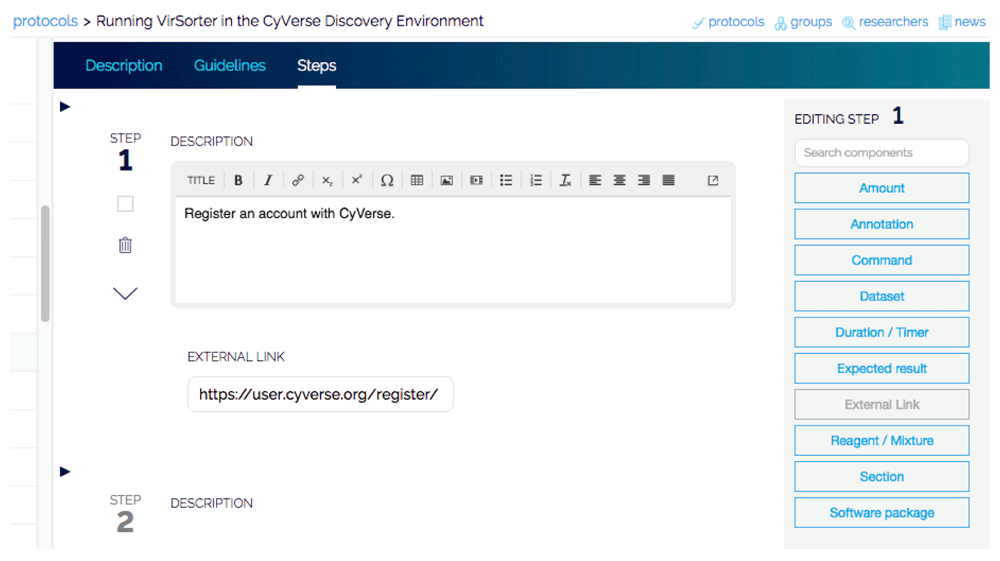
After registration, new protocols can be entered (Figure 1). By default, all protocols are private and can be shared with individual collaborators or any of the groups. The protocols are structured with tabs for the “steps,” “description,” | After registration, new protocols can be entered (Figure 1). By default, all protocols are private and can be shared with individual collaborators or any of the groups. The protocols are structured with tabs for the “steps,” “description,” “guidelines,” and “comments.” When entering the steps, a list of components that can be added to the steps is located on the far right and allows a clear detailing of wetlab or computational portions of the method. Related steps of the protocol can also be easily grouped together into sections such as "preparation," "DNA extraction," and "analysis," etc. Steps may be entered one by one by typing into the text box or by pasting steps from another file, facilitating import of existing protocols. For each step, annotations can be added to make notes on specific steps. Once complete, the protocol can be run in a step-by-step format. | ||
[[File:Fig1 Kindler F1000Res2017 5.gif|800px]] | |||
{{clear}} | |||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| border="0" cellpadding="5" cellspacing="0" width="800px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 1.''' Entering a protocol in protocols.io. Protocols are entered by providing a broad description, information about authors, any prior materials or background required, and detailed step-by-step methods to implement the protocol. Protocols can remain private to an individual or group, or released to the public.</blockquote> | |||
|- | |||
|} | |||
|} | |||
Once a protocol has been created, there are several options for sharing it with collaborators or a group. To make the protocol publicly viewable, one will need to click the "publish" button. A protocol can be reassigned to another individual with a protocols.io account. For ongoing development and changes to adding and using protocols, see the tutorials (http://www.protocols.io/help) at protocols.io.<ref name="protoHelp">{{cite web |url=https://www.protocols.io/help/explore |title=Explore protocols.io |publisher=ZappyLab, Inc |accessdate=21 March 2016}}</ref> | |||
===Developing groups in protocols.io=== | |||
To create a group, one must have an account and be logged in. For example, here we describe the VERVE Net group; however, it is possible to create any group. To create a group, users can click on their personal icon in the upper right-hand corner and select “+ new group.” They will be prompted to enter a group name, image, description of the group, research interests, external website address, physical location of the group, and an affiliation. The user will also decide if the group is open to anyone, by invitation only, or open to membership requests. In addition, the user can choose if the group is visible to others or private. Users are able to invite members into their group and control the privileges of their members. Moreover, as the owner of a group, the user is able to invite other subgroups, such as in the VERVE example, where individual labs are subgroups. | |||
===Finding protocols=== | |||
Protocols on protocols.io can be tagged to allow users to quickly find protocols or collections of protocols in a particular area of interest. Users can also find protocols or other content using the global search at the top of each page, allowing users to search within the entire forum or specific sections of the forum. | |||
===Providing feedback on protocols=== | |||
protocols.io offers three methods for feedback directly from users: twitter, email to protocols developers (info@protocols.io), and through a feedback forum where users and developers alike can respond. These comments are then used to fuel future development. Further, protocols.io recently initiated an ambassadors program where power users (usually graduate students or postdocs) that are directly connected to diverse communities provide feedback from a user perspective. Thus, future development is guided by community input from these sources. | |||
==Use case: VERVE Net: Virus Ecology Research and Virtual Exchange Network== | |||
===Molecular and bioinformatics protocols=== | |||
Often, detailed “tricks of the trade” associated with lab, field, and bioinformatics protocols are not well-described in publications, and at best they are stashed in supplemental materials. Practical information associated with running these protocols under varied conditions cannot be curated, documented, or discussed among students, postdocs, technicians, and faculty working in virology. Moreover, knowledge on when to use a particular version of a given protocol is not easily captured. Protocols.io provides a flexible mechanism wherein protocols can be documented in a step-wise fashion to easily pivot between molecular and bioinformatics methodologies, link to useful websites or code in Github<ref name="DabbishSocial12">{{cite journal |title=Social coding in GitHub: Transparency and collaboration in an open software repository |journal=Proceedings of the ACM 2012 Conference on Computer Supported Cooperative Work |author=Dabbish, L.; Stuart, C.; Tsay, J.; Herbsleb, J. |volume=2012 |pages=1277-1286 |year=2012 |doi=10.1145/2145204.2145396}}</ref>, as well as reference manuals or original source materials for protocols, as exemplified in the VERVENet forum. | |||
The user entering the protocol may not necessarily be the author of the original method. However, by providing links to the primary work, users can attribute credit to the original author while at the same time adding their own updates to the method either while they enter it, or at a later time. Further, other users have the capability to add notes and warnings to existing protocols in protocols.io. This functionality includes a mechanism to email the protocol author for protocol troubleshooting. Corrections and updates made by the protocol authors and users automatically trigger notifications emailed to researchers who use that protocol. Lastly, users can "fork" or copy existing protocols for further refinement or alternate uses while still maintaining links back to the original for credit and reference. As such, the protocol is a living document for the community to reuse and continually refine. | |||
For publication, authors have the option to enter detailed methods into protocols.io, issue a digital object identifier (DOI<ref name="BatesDigital09">{{cite book |title=Encyclopedia of Library and Information Sciences |chapter=Digital Object Identifier (DOI®) System |editor=Bates, M.J.; Maack, M.N. |author=Paskin, N. |publisher=CRC Press |edition=3rd |pages=1586–92 |year=2009 |isbn=9780849397110}}</ref>), and link to the protocols.io record from the methods section. This practice is now being encouraged in journal submissions and by funding agencies. | |||
===Protocol collections=== | |||
Because protocols are often used in conjunction with other protocols, protocols.io has the capability to link protocols into user-defined workflows. This is particularly important for publications that may use a collection of varied protocols (field, lab, and bioinformatics) that are derived from many sources (protocols from the user or other users). In providing a collection of protocols associated with a publication, the authors enable their work to be replicated, easy-to-follow, and transparent to other members of the community in a way that can be referenced and cited. For example, a collection of protocols derived from a recent publication on the human skin double stranded DNA skin virome is available in VERVENet.<ref name="HanniganTheHuman15">{{cite journal |title=The human skin double-stranded DNA virome: Topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome |journal=mBio |author=Hannigan, G.D.; Meisel, J.S.; Tyldsley, A.S. et al. |volume=6 |issue=5 |pages=e01578-15 |year=2015 |doi=10.1128/mBio.01578-15 |pmid=26489866 |pmc=PMC4620475}}</ref><ref name="HanniganTheHuman15_proto">{{cite web |url=https://www.protocols.io/view/The-Human-Skin-dsDNA-Virome-Topographical-and-Temp-ekubcww |title=The human skin double-stranded DNA virome: Topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome |work=protocols.io |author=Hannigan, G.D.; Meisel, J.S.; Tyldsley, A.S. et al. |date=10 March 2016 |doi=10.17504/protocols.io.ekubcww}}</ref> Thus, collections provide a mechanism for furthering open-science efforts. | |||
Protocol collections also provide a mechanism to learn by example for early career scientists or those branching into a new area of scientific inquiry. In particular, detailed protocols associated with a toolkit or workshop — where multimedia options such as slides, video, or links to virtual machines with example datasets and code — can be included.<ref name="HurwitzQIIME15">{{cite web |url=https://www.protocols.io/view/QIIME-Moving-Pictures-of-the-human-microbiome-d5288d |title=QIIME: Moving Pictures of the human microbiome |work=protocols.io |author=Hurwitz, B. |date=24 November 2015 |doi=10.17504/protocols.io.d5288d}}</ref><ref name="CaporasoQIIME10">{{cite journal |title=QIIME allows analysis of high-throughput community sequencing data |journal=Nature Methods |author=Caporaso, J.G.; Kuczynski, J.; Stombaugh, J. et al. |volume=7 |issue=5 |pages=335-6 |year=2010 |doi=10.1038/nmeth.f.303 |pmid=20383131 |pmc=PMC3156573}}</ref> This is particularly important for bioinformatics protocols that often include multiple programs and steps in an analysis for a given publication. Further, individual tools may have a collection of protocols that describe specific use-cases, example datasets, and varied options that they may wish to convey to their users. | |||
==References== | ==References== | ||
Revision as of 20:31, 30 January 2018
| Full article title | Method-centered digital communities on protocols.io for fast-paced scientific innovation |
|---|---|
| Journal | F1000Research |
| Author(s) | Kindler, Lori; Stoliartchouk, Alexei; Teytelman, Leonid; Hurwitz, Bonnie L. |
| Author affiliation(s) | University of Arizona, protocols.io |
| Primary contact | Email: bhurwitz at email dot arizona dot edu |
| Year published | 2017 |
| Volume and issue | 5 |
| Page(s) | 2271 |
| DOI | 10.12688/f1000research.9453.2 |
| ISSN | 2046-1402 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://f1000research.com/articles/5-2271/v2 |
| Download | https://f1000research.com/articles/5-2271/v2/pdf (PDF) |
Abstract
The internet has enabled online social interaction for scientists beyond physical meetings and conferences. Yet despite these innovations in communication, dissemination of methods is often relegated to just academic publishing. Further, these methods remain static, with subsequent advances published elsewhere and unlinked. For communities undergoing fast-paced innovation, researchers need new capabilities to share, obtain feedback, and publish methods at the forefront of scientific development. For example, a renaissance in virology is now underway given the new metagenomic methods to sequence viral DNA directly from an environment. Metagenomics makes it possible to “see” natural viral communities that could not be previously studied through culturing methods. Yet, the knowledge of specialized techniques for the production and analysis of viral metagenomes remains in a subset of labs. This problem is common to any community using and developing emerging technologies and techniques. We developed new capabilities to create virtual communities in protocols.io, an open-access platform for disseminating protocols and knowledge at the forefront of scientific development. To demonstrate these capabilities, we present a virology community forum called VERVENet. These new features allow virology researchers to share protocols and their annotations and optimizations; connect with the broader virtual community to share knowledge, job postings, conference announcements through a common online forum; and discover the current literature through personalized recommendations to promote discussion of cutting edge research. Virtual communities in protocols.io enhance a researcher’s ability to discuss and share protocols, connect with fellow community members, and learn about new and innovative research in the field. The web-based software for developing virtual communities is free to use on protocols.io. Data are available through public APIs at protocols.io.
Introduction
The internet has enabled online social interaction for scientists beyond physical meetings and conferences. Twitter, Facebook, and ResearchGate[1][2][3] provide valuable online forums that many researchers use to share knowledge. At the same time, academic publishing remains time consuming and inefficient for communicating methodology. Protocols are often relegated to supplementary information, if shared at all. There is no good mechanism for easily discussing, troubleshooting, and improving published or unpublished techniques.
This need is even more apparent in emerging fields such as viral ecology where laboratory, field, and bioinformatics methods are being actively developed.[4] For example, new metagenomic techniques to sequence viral DNA directly from environmental samples has led to rapid advances in both molecular and bioinformatic protocols.[5] These protocols, however, are highly specialized and generally used in a few highly proficient labs because: (i) viral metagenomes (viromes) are difficult to produce due to low quantities of DNA and refined isolation and purification methods, (ii) the vast majority of viral sequences are unknown (usually >90%[6]) complicating bioinformatics analyses, and (iii) newly emerging comparative and functional metagenomic analyses exist but require on-going community refinement and development.
Given the experimental nature of methods, the virology community has expressed a need to foster discussions about these protocols towards improved methodologies and increasing connectivity and collaboration among researchers.[7] The challenge is to develop a method-centered collaborative platform that recapitulates the functionality of a scientific meeting - a digital community for connecting with fellow researchers to share and discover state-of-the-art knowledge.
Here we describe new capabilities in protocols.io (http://www.protocols.io), an open-access platform, to create virtual communities for disseminating protocols and knowledge at the forefront of scientific development. To demonstrate these capabilities, we describe a viral ecology community forum called VERVENet (https://www.protocols.io/groups/verve-net) that strives to increase connectivity and knowledge dissemination in viral ecology research at all levels, from undergraduates to accomplished viral ecologists. These new community features enhance a researcher’s ability to discuss and share protocols, connect with fellow community members, and learn about new and innovative research in the field. The web-based software for developing virtual communities is free for use on protocols.io and is further described here.
Protocols.io: A platform to enable methods discussion and dissemination
Protocols.io is a free service for industry and academic scientists to share or maintain private protocols for research.[8] The driving force behind software development is to provide a mechanism for scientists to share improvements and corrections to protocols so that others are not continuously re-discovering knowledge that scientists have not had the time or wear-with-all to publish. Protocols.io provides a free, up-to-date, crowd-sourced protocol repository for the life science community. This software is available as a web-based platform or smart phone application[9][10] to enable mobile solutions for research and bench work. Per best practices in mobile computing, these apps offer extensive options and control of push notifications. In fall 2014, protocols.io offered a well-developed platform for users to share molecular methods; however, no capabilities were in place to share bioinformatics and other methods among groups. To this end, the viral ecology community teamed up with protocols.io to create new group capabilities, develop bioinformatics protocols, and enhance discussion forums for news, methods, and literature.
Introducing VERVENet: The Viral Ecology Research and Virtual Exchange Network
The Viral Ecology Research and Virtual Exchange Network (VERVENet) is a collaboration between the University of Arizona and protocols.io to deliver an online forum for the virology community. To enable this forum, new group functionality was built into protocols.io to promote scientific communication and collaboration. Specifically, group features were developed on top of existing capabilities to share molecular methods in order to (i) share protocols and their annotations and optimizations; (ii) fuel connectivity among viral ecology researchers for sharing data sets, knowledge, job postings, and conference announcements through a common online forum called VERVENet; and (iii) facilitate literature discovery through personalized recommendations to promote discussion on cutting edge viral ecology research. Through developing these interconnected resources in protocols.io for virtual communities, we developed a “go-to” site for viral ecology research.[11] Moreover, these tools are broadly useful to any community or individual lab for promoting scientific inquiry, reproduction of results, dissemination of protocols, and re-use. Specifically, new forums can be created in a matter of minutes to enable connectivity among groups of any size, with tools described here under use cases. The VERVE Net forum is a place to discuss newly emerging methods in viral ecology for any kind of data such as omics or image datasets. However, while images, videos, and tables can be added to protocols/steps to enhance the description of methods, the protocols.io platform is not a data storage site.
Methods
Creating a user profile in protocols.io
Users can view protocols and all public content anonymously, but to interact with the platform, registration is necessary. Registration is quick, as only email and password are required to create an account; however, users are encouraged to create profiles containing their name, website, affiliation, and research interests. Others can search and find a user based on name or keywords. Moreover, user profiles are attached to any material on protocols.io that the user posts publicly. User profiles also contain a field for ORCID[12] so that researchers can tie their profile back to a common identifier and highlight their work in the field. Researchers can also include a biography that describes how they got into the field and what intrigues them. Thus, profiles allow users to add in their own content, rather than simply browse existing content.
Adding protocols in protocols.io
After registration, new protocols can be entered (Figure 1). By default, all protocols are private and can be shared with individual collaborators or any of the groups. The protocols are structured with tabs for the “steps,” “description,” “guidelines,” and “comments.” When entering the steps, a list of components that can be added to the steps is located on the far right and allows a clear detailing of wetlab or computational portions of the method. Related steps of the protocol can also be easily grouped together into sections such as "preparation," "DNA extraction," and "analysis," etc. Steps may be entered one by one by typing into the text box or by pasting steps from another file, facilitating import of existing protocols. For each step, annotations can be added to make notes on specific steps. Once complete, the protocol can be run in a step-by-step format.
|
Once a protocol has been created, there are several options for sharing it with collaborators or a group. To make the protocol publicly viewable, one will need to click the "publish" button. A protocol can be reassigned to another individual with a protocols.io account. For ongoing development and changes to adding and using protocols, see the tutorials (http://www.protocols.io/help) at protocols.io.[13]
Developing groups in protocols.io
To create a group, one must have an account and be logged in. For example, here we describe the VERVE Net group; however, it is possible to create any group. To create a group, users can click on their personal icon in the upper right-hand corner and select “+ new group.” They will be prompted to enter a group name, image, description of the group, research interests, external website address, physical location of the group, and an affiliation. The user will also decide if the group is open to anyone, by invitation only, or open to membership requests. In addition, the user can choose if the group is visible to others or private. Users are able to invite members into their group and control the privileges of their members. Moreover, as the owner of a group, the user is able to invite other subgroups, such as in the VERVE example, where individual labs are subgroups.
Finding protocols
Protocols on protocols.io can be tagged to allow users to quickly find protocols or collections of protocols in a particular area of interest. Users can also find protocols or other content using the global search at the top of each page, allowing users to search within the entire forum or specific sections of the forum.
Providing feedback on protocols
protocols.io offers three methods for feedback directly from users: twitter, email to protocols developers (info@protocols.io), and through a feedback forum where users and developers alike can respond. These comments are then used to fuel future development. Further, protocols.io recently initiated an ambassadors program where power users (usually graduate students or postdocs) that are directly connected to diverse communities provide feedback from a user perspective. Thus, future development is guided by community input from these sources.
Use case: VERVE Net: Virus Ecology Research and Virtual Exchange Network
Molecular and bioinformatics protocols
Often, detailed “tricks of the trade” associated with lab, field, and bioinformatics protocols are not well-described in publications, and at best they are stashed in supplemental materials. Practical information associated with running these protocols under varied conditions cannot be curated, documented, or discussed among students, postdocs, technicians, and faculty working in virology. Moreover, knowledge on when to use a particular version of a given protocol is not easily captured. Protocols.io provides a flexible mechanism wherein protocols can be documented in a step-wise fashion to easily pivot between molecular and bioinformatics methodologies, link to useful websites or code in Github[14], as well as reference manuals or original source materials for protocols, as exemplified in the VERVENet forum.
The user entering the protocol may not necessarily be the author of the original method. However, by providing links to the primary work, users can attribute credit to the original author while at the same time adding their own updates to the method either while they enter it, or at a later time. Further, other users have the capability to add notes and warnings to existing protocols in protocols.io. This functionality includes a mechanism to email the protocol author for protocol troubleshooting. Corrections and updates made by the protocol authors and users automatically trigger notifications emailed to researchers who use that protocol. Lastly, users can "fork" or copy existing protocols for further refinement or alternate uses while still maintaining links back to the original for credit and reference. As such, the protocol is a living document for the community to reuse and continually refine.
For publication, authors have the option to enter detailed methods into protocols.io, issue a digital object identifier (DOI[15]), and link to the protocols.io record from the methods section. This practice is now being encouraged in journal submissions and by funding agencies.
Protocol collections
Because protocols are often used in conjunction with other protocols, protocols.io has the capability to link protocols into user-defined workflows. This is particularly important for publications that may use a collection of varied protocols (field, lab, and bioinformatics) that are derived from many sources (protocols from the user or other users). In providing a collection of protocols associated with a publication, the authors enable their work to be replicated, easy-to-follow, and transparent to other members of the community in a way that can be referenced and cited. For example, a collection of protocols derived from a recent publication on the human skin double stranded DNA skin virome is available in VERVENet.[16][17] Thus, collections provide a mechanism for furthering open-science efforts.
Protocol collections also provide a mechanism to learn by example for early career scientists or those branching into a new area of scientific inquiry. In particular, detailed protocols associated with a toolkit or workshop — where multimedia options such as slides, video, or links to virtual machines with example datasets and code — can be included.[18][19] This is particularly important for bioinformatics protocols that often include multiple programs and steps in an analysis for a given publication. Further, individual tools may have a collection of protocols that describe specific use-cases, example datasets, and varied options that they may wish to convey to their users.
References
- ↑ Ellison, N.B.; Steinfield, C.; Lampe, C. (2007). "The Benefits of Facebook “Friends:” Social Capital and College Students’ Use of Online Social Network Sites". Journal of Computer-Mediated Communication 12 (4): 1143–1168. doi:10.1111/j.1083-6101.2007.00367.x.
- ↑ Kwak, H.; Lee, C.; Park, H.; Moon, S. (2010). "What is Twitter, a social network or a news media?". Proceedings of the 19th International Conference on World Wide Web 2010: 591-600. doi:10.1145/1772690.1772751.
- ↑ Thelwall, M.; Kousha, K. (2015). "ResearchGate: Disseminating, communicating, and measuring Scholarship?". Journal of the Association for Information Science and Technology 66 (5): 876–889. doi:10.1002/asi.23236.
- ↑ Weinbauer, M.G.; Rowe, J.M.; Wilhelm, S.W., ed. (2010). Manual of Aquatic Viral Ecology. American Society of Limnology and Oceanography. doi:10.4319/mave.2010.978-0-9845591-0-7.
- ↑ Brum, J.R.; Sullivan, M.B. (2015). "Rising to the challenge: Accelerated pace of discovery transforms marine virology". Nature Reviews Microbiology 13 (3): 147-59. doi:10.1038/nrmicro3404. PMID 25639680.
- ↑ Hurwitz, B.L.; Sullivan, M.B. (2013). "The Pacific Ocean virome (POV): A marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology". PLoS One 8 (2): e57355. doi:10.1371/journal.pone.0057355. PMC PMC3585363. PMID 23468974. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3585363.
- ↑ "Aquatic Viruses". Facebook. 2 July 2014. https://www.facebook.com/AquaticViruses/posts/760704383968498. Retrieved 09 August 2016.
- ↑ Teytelman, L.; Stoliartchouk, A. (2015). "Protocols.io: Reducing the knowledge that perishes because we do not publish it". Information Services & Use 35 (1–2): 109–115. doi:10.3233/ISU-150769.
- ↑ ZappyLab. "protocols.io". App Store. https://itunes.apple.com/us/app/protocols.io/id976303827. Retrieved 21 March 2016.
- ↑ ZappyLab. "protocols.io". Google Play. https://play.google.com/store/apps/details?id=com.zappylab.protocols. Retrieved 21 March 2016.
- ↑ "VERVE Net". protocols.io. https://www.protocols.io/g/verve-net. Retrieved 21 March 2016.
- ↑ Haak, L.L.; Fenner, M.; Paglione, L. et al. (2012). "ORCID: A system to uniquely identify researchers". Learned Publishing 25 (4): 259–264. doi:10.1087/20120404.
- ↑ "Explore protocols.io". ZappyLab, Inc. https://www.protocols.io/help/explore. Retrieved 21 March 2016.
- ↑ Dabbish, L.; Stuart, C.; Tsay, J.; Herbsleb, J. (2012). "Social coding in GitHub: Transparency and collaboration in an open software repository". Proceedings of the ACM 2012 Conference on Computer Supported Cooperative Work 2012: 1277-1286. doi:10.1145/2145204.2145396.
- ↑ Paskin, N. (2009). "Digital Object Identifier (DOI®) System". In Bates, M.J.; Maack, M.N.. Encyclopedia of Library and Information Sciences (3rd ed.). CRC Press. pp. 1586–92. ISBN 9780849397110.
- ↑ Hannigan, G.D.; Meisel, J.S.; Tyldsley, A.S. et al. (2015). "The human skin double-stranded DNA virome: Topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome". mBio 6 (5): e01578-15. doi:10.1128/mBio.01578-15. PMC PMC4620475. PMID 26489866. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4620475.
- ↑ Hannigan, G.D.; Meisel, J.S.; Tyldsley, A.S. et al. (10 March 2016). "The human skin double-stranded DNA virome: Topographical and temporal diversity, genetic enrichment, and dynamic associations with the host microbiome". protocols.io. doi:10.17504/protocols.io.ekubcww. https://www.protocols.io/view/The-Human-Skin-dsDNA-Virome-Topographical-and-Temp-ekubcww.
- ↑ Hurwitz, B. (24 November 2015). "QIIME: Moving Pictures of the human microbiome". protocols.io. doi:10.17504/protocols.io.d5288d. https://www.protocols.io/view/QIIME-Moving-Pictures-of-the-human-microbiome-d5288d.
- ↑ Caporaso, J.G.; Kuczynski, J.; Stombaugh, J. et al. (2010). "QIIME allows analysis of high-throughput community sequencing data". Nature Methods 7 (5): 335-6. doi:10.1038/nmeth.f.303. PMC PMC3156573. PMID 20383131. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3156573.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added.