Journal:Methods for specifying scientific data standards and modeling relationships with applications to neuroscience
| Full article title | Methods for specifying scientific data standards and modeling relationships with applications to neuroscience |
|---|---|
| Journal | Frontiers in Neuroinformatics |
| Author(s) | Rübel, Oliver; Dougherty, Max; Prabhat; Denes, Peter; Conant, David; Chang, Edward F.; Bouchard, Kristofer |
| Author affiliation(s) | Lawrence Berkeley National Laboratory; University of California, San Francisco |
| Primary contact | Email: oruebel at lbl dot gov -or- kebouchard at lbl dot gov |
| Editors | Ghosh, Satrajit S. |
| Year published | 2016 |
| Volume and issue | 10 |
| Page(s) | 48 |
| DOI | 10.3389/fninf.2016.00048 |
| ISSN | 1662-5196 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | http://journal.frontiersin.org/article/10.3389/fninf.2016.00048/full |
| Download | http://journal.frontiersin.org/article/10.3389/fninf.2016.00048/pdf (PDF) |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
Neuroscience continues to experience a tremendous growth in data; in terms of the volume and variety of data, the velocity at which data is acquired, and in turn the veracity of data. These challenges are a serious impediment to sharing of data, analyses, and tools within and across labs. Here, we introduce BRAINformat, a novel data standardization framework for the design and management of scientific data formats. The BRAINformat library defines application-independent design concepts and modules that together create a general framework for standardization of scientific data. We describe the formal specification of scientific data standards, which facilitates sharing and verification of data and formats. We introduce the concept of "managed objects," enabling semantic components of data formats to be specified as self-contained units, supporting modular and reusable design of data format components and file storage. We also introduce the novel concept of "relationship attributes" for modeling and use of semantic relationships between data objects. Based on these concepts we demonstrate the application of our framework to design and implement a standard format for electrophysiology data and show how data standardization and relationship-modeling facilitate data analysis and sharing. The format uses HDF5, enabling portable, scalable, and self-describing data storage and integration with modern high-performance computing for data-driven discovery. The BRAINformat library is open-source, easy-to-use, and provides detailed user and developer documentation and is freely available at https://bitbucket.org/oruebel/brainformat.
Introduction
Neuroscience research is facing an increasingly challenging data problem due to the growing complexity of experiments and the volume/variety of data being collected from many acquisition modalities. Neuroscientists are routinely collecting data in a broad range of formats that are often highly domain-specific, ad-hoc and/or designed for efficiency with respect to very specific tools and data types. Even for single experiments, scientists are interacting with often tens of different formats — one for each recording device and/or analysis — while many formats are not well-described or are only accessible via proprietary software. Navigating this quagmire of formats hinders efficient analysis, data sharing, and collaboration and can lead to errors and misinterpretations. File formats and standards that can represent neuroscience data and make the data easily accessible play a key role in enabling scientific discovery, development of reusable tools for analysis, and progress toward fostering collaboration in the neuroscience community.
The requirements toward a data format standard for neuroscience are highly complex and go far beyond the needs of traditional, modality-specific formats (e.g., image, audio, or video formats). A neuroscience data format needs to support the management and organization of large collections of data from many modalities and sources such as neurological recordings, external stimuli, recordings of external responses and events (e.g., motion-tracking, video, audio, etc.), derived analytic results, and many others. To enable data interpretation and analysis, the format needs to also support storage of complex metadata, such as descriptions of recording devices, experiments, or subjects, among others.
In addition, a usable and sustainable neuroscience data format needs to satisfy many technical requirements. For example, the format should be self-describing, easy-to-use, efficient, portable, scalable, verifiable, extensible, easy-to-share, and support self-contained and modular storage. Meeting all these complex needs is a daunting challenge. Arguably, the focus of a neuroscience data standard should be on addressing the application-centric needs of organizing scientific data and metadata, rather than on reinventing file storage methods. We here focus on the design of a framework for standardization of data formats while utilizing HDF5 as the underlying data model and storage format. Using HDF5 has the advantage that it already satisfies most of the basic, technical format requirements; HDF5 is self-describing, portable, extensible, widely supported by programming languages and analysis tools, and is optimized for storage and I/O of large-scale scientific data.
In this manuscript we introduce BRAINformat, a novel data format standardization framework and API for scientific data, developed at the Lawrence Berkeley National Labs in collaboration with neuroscientists at the University of California, Berkeley and the University of California, San Francisco. BRAINformat supports the formal specification and verification of scientific data formats and supports the organization of data in a modular, extensible, and reusable fashion via the concept of managed objects (Section 3.1). We introduce the novel concept or relationship attributes for modeling of direct relationships between data objects. Relationship attributes support the specification of structural and semantic links between data, enabling users and developers to formally document and utilize object-to-object relationships in a well-structured and programmatic fashion (Section 3.2). We demonstrate the use of chains of object-to-object relationships to model complex relationships between multi-dimensional arrays based on data registration via the concept of advanced index map relationships (Section 3.2.4). We demonstrate the application of our framework to design and implement a standard format for electrophysiology data and show how data standardization and relationship-modeling facilitate multi-modal data analysis and data sharing (Section 4).
The scientific community utilizes a broad range of data formats. Basic formats explicitly specify how data is laid out and formatted in binary or text data files (e.g., CSV, BOF, etc). While such basic formats are common, they generally suffer from a lack of portability, scalability and a rigorous specification. For text-based files, languages and formats, such as the Extensible Markup Language (XML)[1] or the JavaScript Object Notation (JSON)[2], have become popular means to standardize documents for data exchange. XML, JSON and other text-based standards (in combination with character-encoding schema, e.g., ASCII or Unicode) play a critical role in practice in the exchange of usually relatively small, structured documents but are impractical for storage and exchange of large scientific data arrays.
For storage of large scientific data, HDF5[3] and NetCDF[4] among others, have gained wide popularity. HDF5 is a data model, library, and file format for storing and managing large and complex data. HDF5 supports groups, datasets, and attributes as core data object primitives, which in combination provide the foundation for data organization and storage. HDF5 is portable, scalable, self-describing, and extensible and is widely supported across programming languages and systems, e.g., R, Matlab, Python, C, Fortran, VisIt, or ParaView. The HDF5 technology suite includes tools for managing, manipulating, viewing, and analyzing HDF5 files. HDF5 has been adopted as a base format across a broad range of application sciences, ranging from physics to bio-sciences and beyond.[5] Self-describing formats address the critical need for standardized storage and exchange of complex and large scientific data.
Self-describing formats like HDF5 provide general capabilities for organizing data, but they do not prescribe a data organization. The structure, layout, names, and descriptions of storage objects, hence, often still differ greatly between applications and experiments. This diversity makes the development of common and reusable tools challenging. VizSchema[6] and XDMF[7] among others, propose to bridge this gap between general-purpose, self-describing formats and the need for standardized tools via additional lightweight, low-level schema (often based on XML) to further standardize the description of the low-level data organization to facilitate data exchange and tool development.
Application-oriented formats then generally focus on specifying the organization of data in a semantically meaningful fashion, including but not limited to the specification of storage object names, locations, and descriptions. Many application formats build on existing self-describing formats, e.g., NeXus[8] (neutron, x-ray, and muon data), OpenMSI[9] (mass spectrometry imaging), CXIDB[10] (coherent x-ray imaging), or NetCDF[4] in combination with CF and COARDS metadata conventions for climate data, and many others. Application formats are commonly described by documents specifying the location and names of data items and often provide application-programmer interfaces (API) to facilitate reading and writing of format files. Some formats are further governed by formal, computer-readable, and verifiable specifications. For example, NeXus uses the NXDL[11] XML-based format and schema to define the nomenclature and arrangement of information in a NeXus data file. On the level of HDF5 groups, NeXus also uses the notion of "classes" to define the fields that a group should contain in a reusable and extensible fashion.
The critical need for data standards in neuroscience has been recognized by several efforts over the course of the last several years[12]; however, much work remains. Here, our goal is to contribute to this discussion by providing much-needed methods and tools for the effective design of sustainable neuroscience data standards and demonstration of the methods in practice toward the design and implementation of a usable and extensible format with an initial focus on electrocardiography data. The developers of the KlustaKwik suite[13] have proposed an HDF5-based data format for storage of spike sorting data. Orca (also called BORG)[14] is an HDF5-based format developed by the Allen Institute for Brain Science designed to store electrophysiology and optophysiology data. The NIX project[15] has developed a set of standardized methods and models for storing electrophysiology and other neuroscience data together with their metadata in one common file format based on HDF5. Rather than an application-specific format, NIX defines highly generic models for data as well as for metadata that can be linked to terminologies (defined via odML) to provide a domain-specific context for elements. The open metadata Markup Language odML[16] is a metadata markup language based on XML with the goal to define and establish an open and flexible format to transport neuroscience metadata. NeuroML[17] is also an XML-based format with a particular focus on defining and exchanging descriptions of neuronal cell and network models. The Neurodata Without Borders (NWB)[18] initiative is a recent project with the specific goal "[…] to produce a unified data format for cellular-based neurophysiology data based on representative use cases initially from four laboratories — the Buzsaki group at NYU, the Svoboda group at Janelia Farm, the Meister group at Caltech, and the Allen Institute for Brain Science in Seattle." Members of the NIX, KWIK, Orca, BRAINformat, and other development teams have been invited and contributed to the NWB effort. NWB has adopted concepts and methods from a range of these formats, including from the here-described BRAINformat.
Standardizing scientific data
Data organization and file format API
BRAINformat adopts HDF5 as its main storage backend. HDF5 provides the following primary storage primitives to organize data within HDF5 files:
- Group: A group is used — similar to a folder on a file system — to group zero or more storage objects.
- Dataset: A dataset defines a multidimensional array of data elements, together with supporting metadata (e.g., shape and data type of the array).
- Attribute: Attributes are small datasets that are attached to primary data objects (i.e., groups or datasets) and are used to store additional metadata to further describe the corresponding data object.
- Dimension scale: This is a derived primitive that uses a combination of datasets and attributes to associate datasets with the dimension of another dataset. Dimension scales are used to further characterize dataset dimensions by describing, for example, the time when samples were measured.
Beyond these basic data primitives, we introduce:
- Relationship attributes: Relationship attributes are a novel, custom attribute-type storage primitive that allows us to describe and model structural and semantic relationships between primary data objects in a human-readable and computer-interpretable fashion (described later in the "Modeling data relationships" section).
Neuroscience research inherently relies on complex data collections from many modalities and sources. Examples include neural recordings, audio and video, eye- and motion-tracking, task contingencies, stimuli, analysis results, and many others. It is therefore critically important to specify formats in a modular and extensible fashion while enabling users to easily reuse format modules and integrate new ones. The concept of managed objects allows us to address this central challenge in an easy-to-use and scalable fashion.
Managed objects
A managed object is a primary storage object — i.e., file, group, or dataset — with: (i) a formal, self-contained format specification that describes the storage object and its contents (see the next section "Format specification"), (ii) a specific managed type/class, (iii) a human-readable description, and (iv) an optional unique object identifier, e.g., a DOI. In file, these basic managed object descriptors are stored via standardized attributes. Managed object types may be composed ù i.e., a file or group may contain other managed objects — and further specialized through the concept of inheritance, enabling the independent specification and reuse of data format components. The concept of managed objects significantly simplifies the file format specification process by allowing larger formats to be specified in an easy-to-manage iterative manner. By encapsulating semantic sub-components, managed objects provide an ideal foundation for interacting with data in a manner that is semantically meaningful to applications.
The BRAINformat library provides dedicated base classes to assist with the specification and development of interfaces for new managed object types. The ManagedObject base API implements common features and interfaces to:
- define the specification of a given managed type;
- recursively construct the complete format specification, while automatically resolving nesting of managed objects;
- verify format compliance of a given HDF5 object;
- access common managed object descriptors, e.g., type, description, specification, and object identifier;
- access contained objects, e.g., datasets, groups, managed objects, etc.;
- retrieve all managed objects of a given managed type;
- automatically create manager class instances for HDF5 objects based on their managed type; and
- create new instances of managed objects via a common create(..) method.
To implement a new managed object type, a developer simply defines a new class that inherits from the appropriate base type, i.e., ManagedFile, ManagedGroup, or ManagedDataset. Next, the developer implements the class method get_format_specification(…) to create a formal format specification (see next section) and implements the object method populate(…) to define the type-specific population of managed storage objects to ensure format compliance upon creation, i.e., the goal is to avoid that managed objects can be created in an invalid state to ensure format compliance throughout their life cycle.
BRAINformat efficiently supports self-contained, modular, and mixed data storage strategies, by allowing managed groups and datasets to be stored either directly within the same file as the parent group or separately in an external HDF5 file and included in the parent via an external link. To transparently support external storage of managed objects, we provide a generic file storage container for managed objects. This strategy enables users to create and interact with managed objects in the same manner independent of whether they are stored internal or external to the current HDF5 file, effectively hiding the complexity of interacting with possibly large numbers of files. Being able to effectively use self-contained and modular storage strategies is critical for management of neuroscience data due to the diversity and large number of measurements and derived data products that need to be managed and analyzed in conjunction. Self-contained storage eases data sharing, as all data is available in one file. Modular storage then allows us to dynamically link and integrate complex data collections without requiring expensive data copies, eases management of file sizes, and reduces the risk for file corruption by minimizing changes to existing files.
Format specification
To enable the broad application and use of data formats, it is critical that the underlying data standard is easy to interpret by application scientists as well as unambiguously specified for programmatic interpretation and implementation by developers. Therefore, each format component (i.e., managed object type) is described by a formal, self-contained format specification that is computer-interpretable while at the same time including human-readable descriptions of all components.
We generally assume that format specifications are minimal, i.e., all file objects that are defined in the specification must adhere to the specification, but a user may add custom objects to a file without violating format compliance. The relaxed assumption of a minimal specification ensures on the one hand that we can share and interact with all format-compliant files and components in a standardized fashion, while at the same time enabling users to easily integrate dynamic and custom data (e.g., instrument-specific metadata). This strategy allows researchers to save all their data using standard format components, even if they only partially cover the specific use-case. This is critical to enable scientists to easily adopt file standards and to allow standards to adapt to the ever-evolving experiments, methods, and use-cases in neuroscience and facilitate new science rather than impeding it.
The BRAINformat library defines format specification document standards for files, groups, datasets, attributes, dimensions scales, managed objects, and relationship attributes. All specification documents are based on hierarchical Python dictionaries that can be serialized as JSON documents for persistent storage and sharing. For all data objects we specify the name and/or prefix of the object, whether the object is optional or required, and provide a human-readable textual description of the purpose and content of the object. Depending on the object type (e.g., file, group, dataset, or attribute) additional information is specified, e.g., (i) the datasets, groups, and managed objects contained in a group or file, (ii) attributes for datasets, groups and files, (iii) dimension scales of datasets, (iv) whether a dataset is a primary dataset for visualization and analysis or (v) relationships between objects. Figure 1 shows as an example an abbreviated summary of the format specification of our example electrophysiology data standard described in Section 4 and Supplement 1. Relationship attributes and their specification are discussed further in the "Modeling data relationships" section.
|
The BRAINformat library implements a series of dedicated data structures to facilitate development of format specifications, to ensure their validity, and to simplify the use and interaction with format documents. Using these data structures enables the incremental creation of format specifications, allowing the developer to step-by-step define and compose format specifications. The process of creating new format specifications is in this way much like creating an HDF5 file, making the overall process easily accessible to new users. For example, the following simple code can be used to generate the parts of the BrainDataEphys specification shown in Figure 1:
>>> from brain.dataformat.spec import *
>>> # Define the group
>>> brain_data_ephys = GroupSpec(group=None, prefix=’ephys_data_’, description=“Managed group for storage of raw Ephys recordings.”)
>>> # Define the raw dataset and associated attribute and dimension
>>> raw_data_spec = DatasetSpec(dataset=’raw_data’, prefix=None, optional=False, primary=’True’, description=“Dataset with the Ephys recordings data”)
>>> raw_data_spec.add_attribute( AttributeSpec(attribute=’unit’, prefix=None, value=’Volt’) )
>>> raw_data_spec.add_dimension( DimensionSpec(name=’space’, unit=’id’, dataset=’electrode_id’, axis=0, description=“Id of the recording electrode”) )
>>> # Add the dataset to the group
>>> brain_data_ephys.add_dataset(raw_data_spec, ’ephys_data’)
Using our specification infrastructure we can easily compile a complete data format specification document that lists all managed object types and their format. The following simple example code compiles the specification document for our neuroscience data format directly from the file format API (see also Section 4):
>>> from brain.dataformat.spec import FormatDocument
>>> import brain.dataformat.brainformat as brainformat
>>> json_spec = FormatDocument.from_api(module_object=brainformat).to_json()
Figure 1 shows an abbreviated summary of the result of the above code. Alternatively, we can also recursively construct the complete specification for a given managed object type, e.g., via:
>>> from brain.dataformat.brainformat import BrainDataFile
>>> from brain.dataformat.spec import *
>>> format_spec = BrainDataFile.get_format_specification_recursive() # Construct the document
>>> file_spec = BaseSpec.from_dict(format_spec) # Verification of the document
>>> json_spec = file_spec.to_json(pretty=True) # Convert the document to JSON
In this case, all references to other managed objects are automatically resolved and their specification embedded in the resulting specification document. While the basic specification for BrainDataFile consists only of ≈11 lines of code (see Supplement 1.1.2), the full, recursive specification contains more than 2170 lines (see Supplement 1.2). Being able to incrementally define format specifications is critical because it allows us to easily extend the format in a modular fashion, define and maintain semantic subcomponents in a self-contained fashion, and avoids hard-to-maintain, monolithic, large documents while still making it easy to create comprehensive specifications documents when necessary.
The ability to compile complete format specification documents directly from data format APIs allows developers to easily integrate new format components (i.e., managed object types) in a self-contained fashion simply by adding a new API class without having to maintain separate format specification documents. Furthermore, this strategy avoids inconsistencies between data format APIs and specification documents since format documents are updated automatically.
The concept of managed objects in combination with the format specification language and API provide an application-independent design concept that allows us to define application-specific formats and modules that are built on best practices.
Modeling data relationships
Neuroscience data analytics often relies on complex structural and semantic relationships between datasets. For example, a scientist may use audio recordings to identify particular speech events during the course of an experiment and in turn needs to locate the corresponding data in an electrocorticography recording dataset to study the neural response to the events. In addition, we often encounter structural relationships in data, for example, when using index arrays or when datasets have been acquired simultaneously and/or are using the same recording device, and many others. To enable efficient analysis, reuse, and sharing of neuroscience data it is critical that we can model the diverse relationships between data objects in a structured fashion to enable human and computer discovery, use, and interpretation of relationships.
Modeling data relationships is not well-supported by traditional data formats; this is typically closer to the domain of scientific databases. In HDF5, we can compose data via HDF5 links (soft and hard) and associate datasets with the dimensions of another dataset via the concept of dimension scales. However, these concepts are limited to very specific types of data links that do not describe the semantics of the relationship. A new general approach is needed to describe more complex structural and semantic links between data objects in HDF5.
Specifying and storing relationships
Here we introduce the novel concept of "relationship attributes" to describe complex semantic relationships between a source object and a target data object in a general and extensible fashion. Relationship attributes are associated with the source object and describe how the source is related to the target data object. The source and target of a relationship may be either a HDF5 group or dataset.
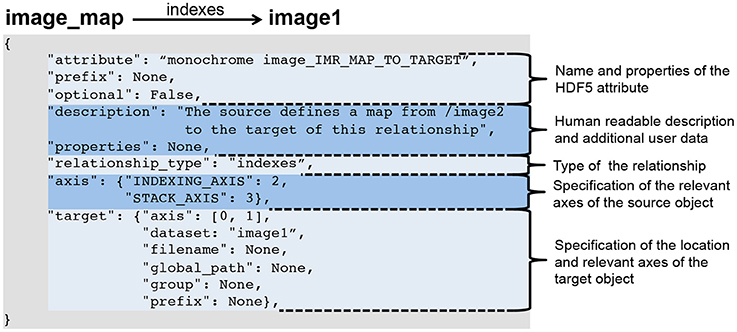
Relationship attributes are like other file components specified via a JSON dictionary and are part of the specification of datasets and groups. Like any other data object, relationships may also be created dynamically to describe relationships that are unknown a priori. Specific instances of relationships are stored as attributes on the source HDF5 object, where the value of the attribute is the JSON document describing the relationship. As illustrated in Figure 2, the JSON specification of a relationship consists of:
- the specification of the name of the attribute and whether the attribute is optional (When stored in HDF5 we prepend the prefix RELATIONSHIP_ATTR_ to the user-defined name of the attribute to describe the attribute's class and ease identification of relationship attributes);
- a human-readable description of the relationship and an optional JSON dictionary with additional user-defined metadata;
- the specification of the type of the relationship (described in the next section);
- the specification of the axes of the source object to which the relationship applies (This may be: (i) a single index, (ii) a list of axes, (iii) a dictionary of axis indices if the axes have a specific user meaning, or iv) none if the relationship applies to the source object as a whole. Note, we do not need to specify the location of the source object, as the specification of the relationship is always associated with either the source object in HDF5 itself or in the format specification.); and
- the specification of the target object describing the location of the object and the axes relevant to the relationship (using the same relative ordering or names of axes as for the source object).
|
Relationship types
The relationship type describes the semantic nature of the relationship. The BRAINformat library currently supports the following relationship types, while additional types can be added in the future:
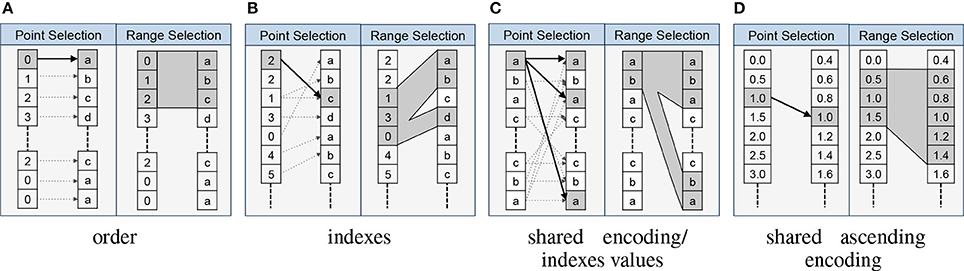
- order: This type indicates that elements along the specified axes of the relationship are ordered in the target in the same way as in the source. This type of relationship is very common in practice. For example, in the case of dimension scales an implicit assumption is that the ordering of elements along the first axis of the scale-dataset matches the ordering of the elements of the dimension it describes. This assumption, however, is only implicit and is by no means always true (nor does HDF5 require this relationship to be true). Using an order relationship we can make this relationship explicit. Other common uses of order relationships include describing the matched ordering of electrodes in datasets that have been recorded using the same device or matched ordering of records in datasets that have been acquired synchronously.
- equivalent: This type expresses that the source and target object encode the same data (even if they might store different values). This relationship type also implies that the source and target contain the same number of values ordered in the same fashion. This relationship occurs in practice any time the same data is stored multiple times with different encodings. For example, to facilitate data processing a user may store a dataset of strings with the names of tokens and store another dataset with the equivalent integer ID of the tokens (e.g., |baa| = 1, |gaa| = 2, etc.).
- indexes: This type describes that the source dataset contains indices into the target dataset or group. In practice this relationship type is used to describe basic data structure where we store, for example, a list of unique values (tokens) along with other arrays that reference that list.
- shared encoding: This type indicates that the source and target data object contain values with the same encoding so that the values can be directly compared via equals “==”. This relationship is useful in practice any time two objects (datasets or groups) contain data with the same encoding (e.g., two datasets describing external stimuli using the same ontology).
- shared ascending encoding: This type implies that the source and target object share the same encoding and that the values are sorted in ascending order in both. The additional constraint on the ordering enables i) comparison of values via greater than “>” and less than “<” in addition to equals == and ii) more efficient processing and comparison of data ranges. For example, in the case of two datasets that encode time, we often find that individual time points do not match exactly between the source and target (e.g., due to different sampling rates). However, due to the ascending ordering of values, a user is still able to compare ranges in time in a meaningful way.
- indexes values: This type is typically used to describe value-based referencing of data and indicates that the source object selects certain parts of the target based on data values (or keys in the case of groups). This relationship is a special type of shared encoding relationship.
- user: The user relationship is a general container to allow users to specify custom semantic relationships that do not match any of the existing relationship patterns. Additional metadata about the relationship may be stored as part of the user-defined properties dictionary of the relationship attribute.
Using relationship attributes
Relationship attributes are a direct extension to the previously described format specification infrastructure. Similar to other main data objects, BRAINformat provides dict-like data structures to help with the formal specification of relationship attributes. In addition, the BRAINformat library also provides a dedicated RelationshipAttribute API, which supports creation and retrieval of relationship attributes (as well as index map relationship, described in Section 3.2.4) and provides easy access to the source and target HDF5 object and corresponding specifications of relationship attributes.
One central advantage of explicitly modeling relationships is that it allows us to formalize the interactions and collaborative usage of related data objects. In particular, the relationship types imply formal rules for how to map data selections from the source to the target of a relationship. The RelationshipAttribute API implements these rules and supports standard data selection operations, which allows us to easily map selections from the source to the target data object using a familiar array syntax. For example, assume we have two datasets A and B that are related via an indexes relationship RA→B. A user now selects the values A[1:10] and wants to locate the corresponding data values in B. Using the BRAINformat API we can now simply write RA→B[1:10] to map the selection [1:10] from the source A to the target B, and if desired retrieve the corresponding data values in B via B[RA→B[1:10]]. Figure 3 provides an overview of the rules for mapping data selections based on the relationship type.
|
References
- ↑ Bray, T.; Paoli, J.; Sperberg-McQueen, C.M. et al. (26 November 2008). "Extensible Markup Language (XML) 1.0 (Fifth Edition)". World Wide Web Consortium. https://www.w3.org/TR/2008/REC-xml-20081126/.
- ↑ Crockford, D.. "Introducing JSON". http://json.org/.
- ↑ "Welcome to the HDF5 Home Page". The HDF Group. https://support.hdfgroup.org/HDF5/.
- ↑ 4.0 4.1 Rew, R.; Davis, G. (1990). "NetCDF: An interface for scientific data access". IEEE Computer Graphics and Applications 10 (4): 76–82. doi:10.1109/38.56302.
- ↑ Habermann, T.; Collette, A.; Vincerna, S. et al. (29 September 2014). "The Hierarchical Data Format (HDF): A Foundation for Sustainable Data and Software". The HDF Group. http://dx.doi.org/10.6084/m9.figshare.1112485.
- ↑ "VizSchema – Visualization interface for scientific data". IADIS International Conference Computer Graphics, Visualization, Computer Vision and Image Processing 2009 2009: 49–56. 2009. ISBN 9789728924843. http://www.iadisportal.org/digital-library/vizschema-%C2%96-visualization-interface-for-scientific-data.
- ↑ "Enhancements to the eXtensible Data Model and Format (XDMF)". DoD High Performance Computing Modernization Program Users Group Conference, 2007 2007. 2007. doi:10.1109/HPCMP-UGC.2007.30. ISBN 9780769530885.
- ↑ Klosowski, P.; Kiennecke, M.; Tischler, J.Z.; Osborn, R. (1997). "NeXus: A common format for the exchange of neutron and synchroton data". Physica B: Condensed Matter 241—243: 151–153. doi:10.1016/S0921-4526(97)00865-X.
- ↑ Rübel, O.; Greiner, A.; Cholia, S. et al. (2013). "OpenMSI: A High-Performance Web-Based Platform for Mass Spectrometry Imaging". Analytical Chemistry 85 (21): 10354–10361. doi:10.1021/ac402540a.
- ↑ Maia, F.R. (2012). "The Coherent X-ray Imaging Data Bank". Nature Methods 9 (9): 854-5. doi:10.1038/nmeth.2110. PMID 22936162.
- ↑ "3.2 NXDL: The NeXus Definition Language". NeXus User Manual and Reference Documentation. NeXus International Advisory Committee. http://download.nexusformat.org/doc/html/nxdl.html.
- ↑ Teeters, J.; Benda, J.; Davison, A. et al. (3 June 2016). "Requirements for storing electrophysiology data". Cornell University Library. https://arxiv.org/abs/1605.07673.
- ↑ Kadir, S.N.; Goodman, D.F.M.; Harris, K.D.. "Kwik format". http://klusta.readthedocs.io/en/latest/kwik/.
- ↑ Godfrey, K. (20 November 2014). "Orca file format for e-phys and o-phys neurodata" (PDF). Neurodata Without Borders Hackathon. http://crcns.org/files/data/nwb/h1/NWBh1_09_Keith_Godfrey.pdf.
- ↑ Stoewer, A.; Kellner, C.J.; Grewe, J. (2016). "Nix: Home". https://github.com/G-Node/nix/wiki.
- ↑ Grewe, J.; Wachtler, T.; Benda, J. (2011). "A bottom-up approach to data annotation in neurophysiology". Frontiers in Neuroinformatics 5: 16. doi:10.3389/fninf.2011.00016.
- ↑ Gleeson, P.; Crook, S.; Cannon, R.C. (2010). "NeuroML: A language for describing data driven models of neurons and networks with a high degree of biological detail". PLoS Computational Biology 6 (6): e1000815. doi:10.1371/journal.pcbi.1000815. PMC PMC2887454. PMID 20585541. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2887454.
- ↑ Teeters, J.L.; Godfrey, K.; Young, R. et al. (2015). "Neurodata Without Borders: Creating a common data format for neurophysiology". Neuron 88 (4): 629–34. doi:10.1016/j.neuron.2015.10.025. PMID 26590340.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. References are in order of appearance rather than alphabetical order (as the original was). The original URL to klusta Kwik format was no longer valid, and an updated URL was used.