Difference between revisions of "Journal:NG6: Integrated next generation sequencing storage and processing environment"
Shawndouglas (talk | contribs) (Created stub. Saving and adding more.) |
Shawndouglas (talk | contribs) (Added content. Saving and adding more later.) |
||
| Line 42: | Line 42: | ||
NG6 is an extensible sequencing provider oriented LIMS. It includes read quality control and first level analysis processes which ease the data validation made jointly by the sequencing facility staff and the end-users. It provides a secured user-friendly interface to visualize and download the raw sequences files and the analysis results. | NG6 is an extensible sequencing provider oriented LIMS. It includes read quality control and first level analysis processes which ease the data validation made jointly by the sequencing facility staff and the end-users. It provides a secured user-friendly interface to visualize and download the raw sequences files and the analysis results. | ||
==Implementation== | |||
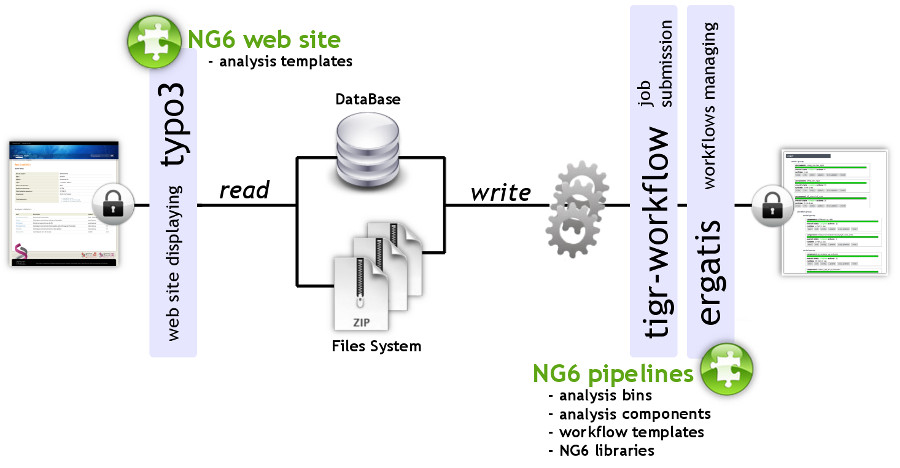
NG6 can be split into two distinct parts: the pipelines and the web site (Figure 1). The pipelines gather a set of analyses adapted to the produced sequences. They can only be accessed and launched by the sequencing facility team. The pipelines are running in Ergatis<ref name="OrvisErgatis10">{{cite journal |title=Ergatis: A web interface and scalable software system for bioinformatics workflows |journal=Bioinformatics |author=Orvis, J.; Crabtree, J.; Galens, K. et al. |volume=26 |issue=12 |pages=1488-1492 |year=2010 |doi=10.1093/bioinformatics/btq167 |pmid=20413634 |pmc=PMC2881353}}</ref>: a workflow management system able to iterate through multiple inputs in order to run them at the same time on a computer farm. These jobs perform analysis and save the analysis results in the NG6 database and directories. The web site part, presenting the results has been implemented as a TYPO3<ref name="TYPO3Home">{{cite web |url=https://typo3.org/ |title=TYPO3 |publisher=TYPO3 Association}}</ref> extension. | |||
[[File:Fig1 Mariette BMCGenomics2015 13.jpg|800px]] | |||
{{clear}} | |||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| border="0" cellpadding="5" cellspacing="0" width="800px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 1. Achitecture of the ng6 application.''' NG6 pipelines are available within the Ergatis workflow environment. The analyses are processed either on a local system or on a distributed environment. While the analyses are running, they store the resulting files on the file system and add information about the run in the database. The produced data is then displayed by the NG6 extension of the TYPO3 CMS. Both NG6 web site and NG6 pipelines are accessible through a web browser after authentication.</blockquote> | |||
|- | |||
|} | |||
|} | |||
NG6 uses three data types: project, run and analysis. A project is a collection of runs and analysis. A run contains one or several raw files which can be used as inputs of different analysis. A project is owned by a user group and only users within this group are allowed to browse and download data related to this project. | |||
===Building and running pipelines=== | |||
Pipelines are defined by a set of connected Ergatis components. Depending on the links between the components, they are processed in a parallel or a serial manner. Most components available in NG6 combine a processing step and a storage step. This last one stores, on one hand, resulting files into the ad-hoc directory structure and, on the other hand, saves information into the database such as software version, parameters, links between analysis and resulting figures. | |||
In the current version, NG6 offers a set of pipelines adapted to two platforms (Roche 454, Illumina HiSeq), four file formats (sff, fastq, fasta and qseq) and handles both Casava 1.7 and Casava 1.8 outputs of the Illumina package.<ref name="IlluminaHome">{{cite web |url=http://www.illumina.com/ |title=Illumina |publisher=Illumina, Inc}}</ref> It includes analyses such as quality control, genomic read alignment, BAC assembly, 16S/18S diversity analysis, expression quantification using 16S amplicons. In order to handle multiplexed runs, some pipelines first split the input read file into sample files, process and collect results on each of them and last merge these results in a summary table. | |||
As an example, the 454_default pipeline processes sff files, coming from the Roche sequencer. It first performs usual statistical analysis on the reads, then tracks down contamination from common contaminant databases (ecoli, yeast and phage) using BLAST<ref name="AltschulBLAST90">{{cite journal |author=Altschul, S.; Gish, W.; Miller, W.; Myers, E.; Lipman, D. |title=Basic local alignment search tool |doi=10.1016/S0022-2836(05)80360-2 |journal=Journal of Molecular Biology |volume=215 |issue=3 |pages=403–410 |year=1990 |pmid=2231712}}</ref> returning a list of contaminated sequence IDs. Contamination between the different regions is also traced using the sfffile script included in the Roche Newbler package.<ref name="Roche454Home">{{cite web |url=http://www.my454.com/ |title=454 Sequencing |publisher=Roche Diagnostics Corporation}}</ref> Sequences with incorrect MID (Multiplexed ID) are discarded and the number of contaminated sequences is returned to the end-user. Roche 454 sequencing kits include control fragments known as spike-ins within each run. Statistics on the corresponding sequences are used to check if the run matches the expected quality standard. In the next step reads are cleaned using the PyroCleaner script.<ref name="MarieteAssess11">{{cite journal |title=Assessment of replicate bias in 454 pyrosequencing and a multi-purpose read-filtering tool |journal=BMC Research Notes |author=Mariette, J.; Noirot, C.; Klopp, C. |volume=4 |pages=149 |year=2011 |doi=10.1186/1756-0500-4-149 |pmid=21615897 |pmc=PMC3117718}}</ref> It discards reads considering different criteria such as length, base quality, complexity, number of undetermined bases, multiple copy reads or even faulty paired-ends. The analysis results are presented to the users in a summary table. Last, a ''de novo'' assembly is performed on the cleaned reads using the Newbler runAssembly command.<ref name="Roche454Home" /> Some basic figures regarding the assembly results, such as contig count, N50 value, contig length distribution or even contig length versus sum of read length per contig diagram are presented to the user in order to ease the assembly quality assessment. | |||
==References== | ==References== | ||
| Line 47: | Line 70: | ||
==Notes== | ==Notes== | ||
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. | This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. Additionally, numerous proper nouns were not capitalized originally but have been updated in this text. | ||
<!--Place all category tags here--> | <!--Place all category tags here--> | ||
Revision as of 02:44, 15 March 2016
| Full article title | NG6: Integrated next generation sequencing storage and processing environment |
|---|---|
| Journal | BMC Genomics |
| Author(s) | Mariette, J.; Escudié, F.; Allias, N.; Salin, G.; Noirot, C.; Thomas, S.; Klopp, C. |
| Author affiliation(s) | Biométrie et Intelligence Artificielle and Génétique Cellulaire |
| Primary contact | E-mail: Jerome.Mariette@toulouse.inra.fr |
| Year published | 2012 |
| Volume and issue | 13 |
| Page(s) | 462 |
| DOI | 10.1186/1471-2164-13-462 |
| ISSN | 1471-2164 |
| Distribution license | Creative Commons Attribution 2.0 Generic |
| Website | http://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-13-462 |
| Download | http://bmcgenomics.biomedcentral.com/track/pdf/10.1186/1471-2164-13-462 (PDF) |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
Background
Next generation sequencing platforms are now well implanted in sequencing centres and some laboratories. Upcoming smaller scale machines such as the 454 junior from Roche or the MiSeq from Illumina will increase the number of laboratories hosting a sequencer. In such a context, it is important to provide these teams with an easily manageable environment to store and process the produced reads.
Results
We describe a user-friendly information system able to manage large sets of sequencing data. It includes, on one hand, a workflow environment already containing pipelines adapted to different input formats (sff, fasta, fastq and qseq), different sequencers (Roche 454, Illumina HiSeq) and various analyses (quality control, assembly, alignment, diversity studies,…) and, on the other hand, a secured web site giving access to the results. The connected user will be able to download raw and processed data and browse through the analysis result statistics. The provided workflows can easily be modified or extended and new ones can be added. Ergatis is used as a workflow building, running and monitoring system. The analyses can be run locally or in a cluster environment using Sun Grid Engine.
Conclusions
NG6 is a complete information system designed to answer the needs of a sequencing platform. It provides a user-friendly interface to process, store and download high-throughput sequencing data.
Background
Sequencer manufacturers follow different objectives using different platforms.[1] In the first place they release upgrades of second generation platforms producing more data with updated hardware and sequencing kits. This lowers the sequencing cost per base pair but often focuses these machines on medium or large projects. In the second place, they introduce new laboratory scale platforms such as the Illumina MiSeq or the Roche Junior which target smaller projects. And last, they work on the third generation machines which will not depend on amplified material and therefore get rid of some biases. The first two machines types which are already marketed today associated with a larger scope of sequencing protocols, enabling new studies, push towards more sequencing projects and more users.
Once the sequencing is done, the largest part of the work and the longest time period of the project are dedicated to data analysis. Therefore it is important to provide the new smaller production units and the laboratories in which the projects are conducted with efficient and user-friendly processing environments, enabling quality control and routine analysis. These pieces of software should have several features such as access control, metadata storage on the produced reads, quality control including known bias verification and standard analysis. NG6 was developed to match these goals and to be as flexible as possible, in order to follow sequencing technologies upgrades.
Laboratory information management systems (LIMS) are often focused on the traceability of the biological material. Some of them, such as PIMS[2] or even SLIMS[3], have included extensions to monitor the sequencing process. However few of the open-source LIMS also provide the data processing environment. This feature is present in the Galaxy[4] sample tracking module. It is based on the Galaxy workflow engine and provides users with an interface to create and track sequencing requests. Once the sequences have been produced, the user can transfer its data files, build and run workflows to process them.
NG6 is an extensible sequencing provider oriented LIMS. It includes read quality control and first level analysis processes which ease the data validation made jointly by the sequencing facility staff and the end-users. It provides a secured user-friendly interface to visualize and download the raw sequences files and the analysis results.
Implementation
NG6 can be split into two distinct parts: the pipelines and the web site (Figure 1). The pipelines gather a set of analyses adapted to the produced sequences. They can only be accessed and launched by the sequencing facility team. The pipelines are running in Ergatis[5]: a workflow management system able to iterate through multiple inputs in order to run them at the same time on a computer farm. These jobs perform analysis and save the analysis results in the NG6 database and directories. The web site part, presenting the results has been implemented as a TYPO3[6] extension.
|
NG6 uses three data types: project, run and analysis. A project is a collection of runs and analysis. A run contains one or several raw files which can be used as inputs of different analysis. A project is owned by a user group and only users within this group are allowed to browse and download data related to this project.
Building and running pipelines
Pipelines are defined by a set of connected Ergatis components. Depending on the links between the components, they are processed in a parallel or a serial manner. Most components available in NG6 combine a processing step and a storage step. This last one stores, on one hand, resulting files into the ad-hoc directory structure and, on the other hand, saves information into the database such as software version, parameters, links between analysis and resulting figures.
In the current version, NG6 offers a set of pipelines adapted to two platforms (Roche 454, Illumina HiSeq), four file formats (sff, fastq, fasta and qseq) and handles both Casava 1.7 and Casava 1.8 outputs of the Illumina package.[7] It includes analyses such as quality control, genomic read alignment, BAC assembly, 16S/18S diversity analysis, expression quantification using 16S amplicons. In order to handle multiplexed runs, some pipelines first split the input read file into sample files, process and collect results on each of them and last merge these results in a summary table.
As an example, the 454_default pipeline processes sff files, coming from the Roche sequencer. It first performs usual statistical analysis on the reads, then tracks down contamination from common contaminant databases (ecoli, yeast and phage) using BLAST[8] returning a list of contaminated sequence IDs. Contamination between the different regions is also traced using the sfffile script included in the Roche Newbler package.[9] Sequences with incorrect MID (Multiplexed ID) are discarded and the number of contaminated sequences is returned to the end-user. Roche 454 sequencing kits include control fragments known as spike-ins within each run. Statistics on the corresponding sequences are used to check if the run matches the expected quality standard. In the next step reads are cleaned using the PyroCleaner script.[10] It discards reads considering different criteria such as length, base quality, complexity, number of undetermined bases, multiple copy reads or even faulty paired-ends. The analysis results are presented to the users in a summary table. Last, a de novo assembly is performed on the cleaned reads using the Newbler runAssembly command.[9] Some basic figures regarding the assembly results, such as contig count, N50 value, contig length distribution or even contig length versus sum of read length per contig diagram are presented to the user in order to ease the assembly quality assessment.
References
- ↑ Glenn, T.C. (2011). "Field guide to next-generation DNA sequencers". Molecular Ecology Resources 11 (5): 759-769. doi:10.1111/j.1755-0998.2011.03024.x. PMID 21592312.
- ↑ Troshin, P.V. Postis, V.L.; Ashworth, D. et al. (2011). "PIMS sequencing extension: a laboratory information management system for DNA sequencing facilities". BMC Research Notes 4: 48. doi:10.1186/1756-0500-4-48. PMC PMC3058032. PMID 21385349. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3058032.
- ↑ Van Rossum, T.; Tripp, B.; Daley, D. (2010). "SLIMS: A user-friendly sample operations and inventory management system for genotyping labs". Bioinformatics 26 (14): 1808-1810. doi:10.1093/bioinformatics/btq271. PMC PMC2894515. PMID 20513665. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2894515.
- ↑ Giardine, B.; Riemer, C.; Hardison, R.C. et al. (2005). "Galaxy: A platform for interactive large-scale genome analysis". Genome Research 15 (10): 1451–1455. doi:10.1101/gr.4086505. PMC PMC1240089. PMID 16169926. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1240089.
- ↑ Orvis, J.; Crabtree, J.; Galens, K. et al. (2010). "Ergatis: A web interface and scalable software system for bioinformatics workflows". Bioinformatics 26 (12): 1488-1492. doi:10.1093/bioinformatics/btq167. PMC PMC2881353. PMID 20413634. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2881353.
- ↑ "TYPO3". TYPO3 Association. https://typo3.org/.
- ↑ "Illumina". Illumina, Inc. http://www.illumina.com/.
- ↑ Altschul, S.; Gish, W.; Miller, W.; Myers, E.; Lipman, D. (1990). "Basic local alignment search tool". Journal of Molecular Biology 215 (3): 403–410. doi:10.1016/S0022-2836(05)80360-2. PMID 2231712.
- ↑ 9.0 9.1 "454 Sequencing". Roche Diagnostics Corporation. http://www.my454.com/.
- ↑ Mariette, J.; Noirot, C.; Klopp, C. (2011). "Assessment of replicate bias in 454 pyrosequencing and a multi-purpose read-filtering tool". BMC Research Notes 4: 149. doi:10.1186/1756-0500-4-149. PMC PMC3117718. PMID 21615897. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3117718.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. Additionally, numerous proper nouns were not capitalized originally but have been updated in this text.