Difference between revisions of "Journal:No specimen left behind: Industrial scale digitization of natural history collections"
Shawndouglas (talk | contribs) (Added content. Saving and adding more.) |
Shawndouglas (talk | contribs) (Added content. Saving and adding more.) |
||
| Line 19: | Line 19: | ||
|download = Click "PDF" button on website to generate | |download = Click "PDF" button on website to generate | ||
}} | }} | ||
{{ombox | |||
| type = content | |||
| style = width: 500px; | |||
| text = This article should not be considered complete until this message box has been removed. This is a work in progress. | |||
}} | |||
==Abstract== | ==Abstract== | ||
Traditional approaches for digitizing natural history collections, which include both imaging and metadata capture, are both labour- and time-intensive. Mass-digitization can only be completed if the resource-intensive steps, such as specimen selection and databasing of associated [[information]], are minimized. Digitization of larger collections should employ an “industrial” approach, using the principles of automation and crowd sourcing, with minimal initial metadata collection including a mandatory persistent identifier. A new workflow for the mass-digitization of natural history museum collections based on these principles, and using SatScan® tray scanning system, is described. | Traditional approaches for digitizing natural history collections, which include both imaging and metadata capture, are both labour- and time-intensive. Mass-digitization can only be completed if the resource-intensive steps, such as specimen selection and databasing of associated [[information]], are minimized. Digitization of larger collections should employ an “industrial” approach, using the principles of automation and crowd sourcing, with minimal initial metadata collection including a mandatory persistent identifier. A new workflow for the mass-digitization of natural history museum collections based on these principles, and using SatScan® tray scanning system, is described. | ||
| Line 168: | Line 172: | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"|39 | | style="background-color:white; padding-left:10px; padding-right:10px;"|39 | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;" rowspan="2"|Canon MP-E65 lens, 1× | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" rowspan="2"|4272×2848 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|2.8 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|0.5 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|5–6 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|57 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|8.8 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|16 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|4 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|- | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" rowspan="2"|Canon MP-E65 lens, 5× | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" rowspan="2"|4272×2848 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|2.8 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|<0.3 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|8–1 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|256 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|2 | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|16 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|2 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|6–2 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|71.8 | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|7 | |||
|- | |||
|} | |||
|} | |||
===Metadata capture=== | |||
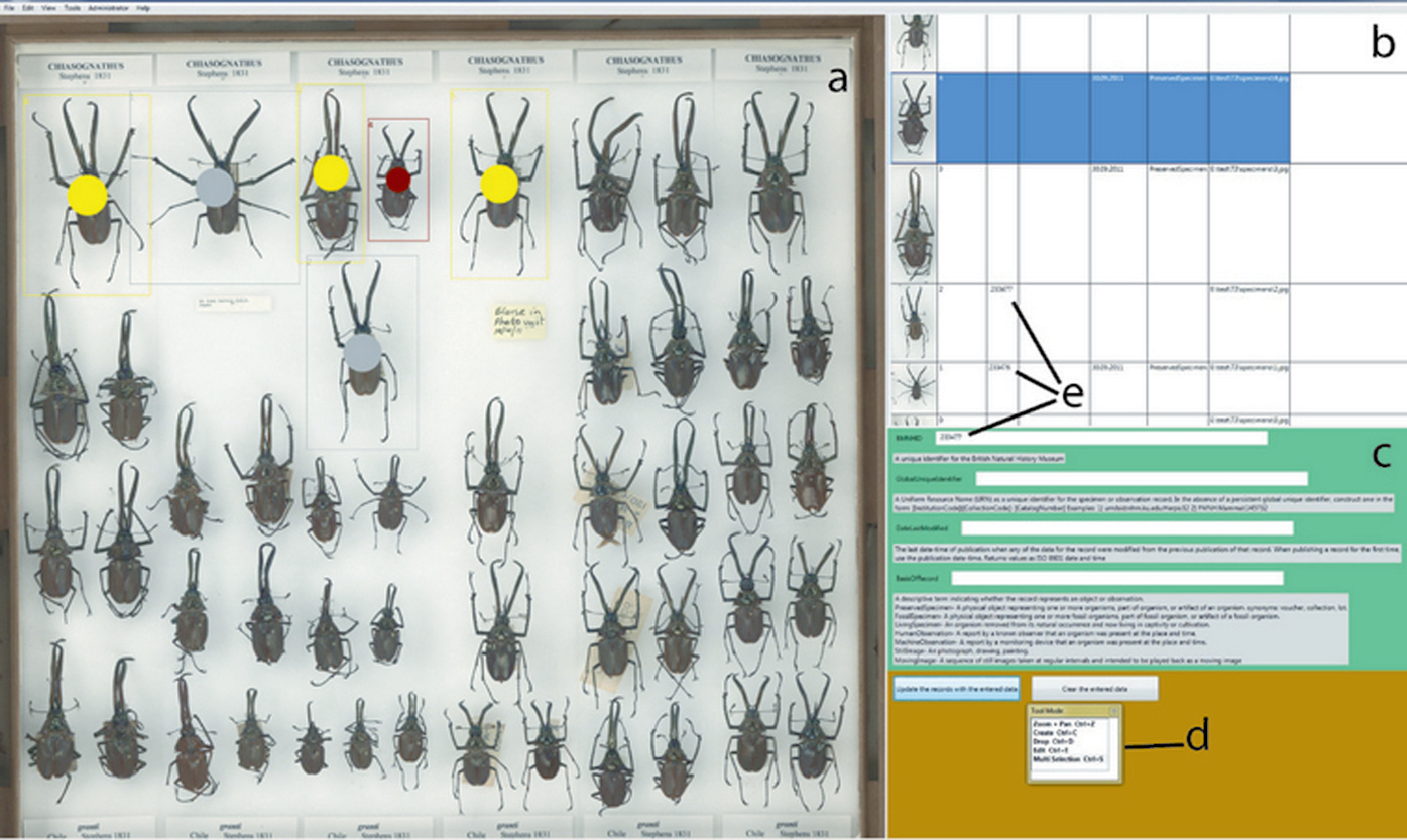
A prototype software program, Metadata Creator, has been designed to allow fast capture of specimen data and associating these with the image of the specimen (Fig. 3). Users can mark individual specimens on the panoramic image by drawing rectangular boxes around them, selecting these areas and annotating them individually or in batches. Methods for marking the specimen, editing regions of interest and selection of multiple specimens are analogous to those used in many common graphic applications and so will be familiar, even to inexperienced users. | |||
Specimen metadata is captured in a series of fields that are compatible with the Darwin Core 1.4.1 schema (http://rs.tdwg.org/dwc/) and which can be customized to particular user requirements. To maximize throughput, only basic metadata are collected at this stage. These will generally include a unique collection number of every specimen (see below, barcodes), collection identification (to the available curatorial level, e.g. to species/subspecies for the “Main Collection” and family/order for unsorted accessions), and, if possible, biogeographic region/country. Taxon names are looked up from an index derived from the NHM Collections Management Database. A completed project comprises a folder with an archival image of the drawer, full-resolution images of individual specimens cut-out from the drawer image, and an XML file containing annotations and links to specimen images (Appendix 1). Trials have demonstrated that 10–20 seconds per specimen is required to capture basic metadata using the Metadata Creator Software. A unique ID for the drawer is also recorded. As the NHM Collection Management System already includes a complete collections index (a brief description of the content of every drawer), no additional information is required. | |||
[[File:Fig3 BlagoderovZooKeys2012 209.jpg|900px]] | |||
{{clear}} | |||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| border="0" cellpadding="5" cellspacing="0" width="900px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 3.''' Metadata Creator software: '''a–c''' working areas '''a''' drawer image '''b''' specimen records '''c''' annotation fields '''d''' tool selector '''e''' unique IDs</blockquote> | |||
|- | |||
|} | |} | ||
|} | |} | ||
===Assigning uIDs=== | |||
Every specimen is assigned a unique number under which it will be registered in the NHM Collections Management Database. It is a requirement of collections management procedures that a label bearing the specimen’s uID is attached to the specimen. To streamline this part of the process, it is subdivided into the following steps: | |||
# A sequence of unique numbers is generated from the NHM Collections Management Database. | |||
# Labels that include both a human-readable number and a machine-readable barcode are printed. | |||
# The operator labels the specimens by selecting a specimen on the drawer image, pinning a label under the specimen, and scanning the barcode, thereby adding the uID into the corresponding field of Metadata Creator. Barcodes can be pinned facing up or down depending on curatorial practice; the former has the advantage of visibility on the image. In this case imaging, of course, has to take place after assigning uIDs. Images of individual specimens for which the metadata have been collected and individual numbers assigned are automatically marked on the drawer image with a grey spot, allowing easy visualization of progress. | |||
# When all specimens have been labelled and recorded, the XML file and corresponding specimen images are imported into the NHM Collections Management Database. | |||
We must emphasize that Metadata Creator is a prototype software application; much more development is needed for to perfect its functionality, user interface, and integration with the Museum’s information systems. | |||
==References== | ==References== | ||
Revision as of 01:06, 4 March 2016
| Full article title | No specimen left behind: Industrial scale digitization of natural history collections |
|---|---|
| Journal | ZooKeys |
| Author(s) | Blagoderov, V.; Kitching, I.J.; Livermore, L.; Simonsen, T.J.; Smith, V.S. |
| Author affiliation(s) | Natural History Museum - London |
| Primary contact | E-mail: v.blagoderov@nhm.ac.uk |
| Editors | Penev, L. |
| Year published | 2012 |
| Volume and issue | 209 |
| Page(s) | 133-146 |
| DOI | 10.3897/zookeys.209.3178 |
| ISSN | 1313-2970 |
| Distribution license | Creative Commons Attribution 3.0 Unported |
| Website | http://zookeys.pensoft.net/articles.php?id=2916 |
| Download | Click "PDF" button on website to generate |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
Traditional approaches for digitizing natural history collections, which include both imaging and metadata capture, are both labour- and time-intensive. Mass-digitization can only be completed if the resource-intensive steps, such as specimen selection and databasing of associated information, are minimized. Digitization of larger collections should employ an “industrial” approach, using the principles of automation and crowd sourcing, with minimal initial metadata collection including a mandatory persistent identifier. A new workflow for the mass-digitization of natural history museum collections based on these principles, and using SatScan® tray scanning system, is described.
Keywords: Digitization, imaging, specimen metadata, natural history collections, biodiversity informatics
Introduction
Natural history collections are of immense scientific and cultural importance. Specimens in public museums and herbaria and their associated data represent a potentially vast repository of information on biodiversity, ecosystems and natural resources for the widest range of stakeholders, from governments and NGOs to schools and private individuals. Numerous examples of the uses to which biodiversity data derived from natural history collections have been put in research on evolution and genetics, nature conservation and resource management, public health and safety, and education are widely available (summarized in Chapman 2005, Baird 2010).[1][2] The universe of natural history collection data has been estimated to be between 1.2 and 2.1 × 109 units (specimens, lots and collections) (Ariño 2010).[3] To ensure efficient access, dissemination and exploitation of such an immense wealth of biodiversity relevant data, it is evident that a well-coordinated and streamlined approach to global digitization is required, in particular because it is absolutely essential for the scientific value of the generated data that the outputs (images, metadata, etc.) are linked together and also back to the original specimens via unique identifiers (uIDs).
In recent years, substantial efforts and resources have been invested into the digitization of natural history collections, with museums and herbaria routinely employing specimen level collection databases to replace older, paper-based card indexes and ledgers. In theory, this should make dissemination of specimen data through biodiversity informatics portals such as the Global Biodiversity Information Facility (GBIF; http://www.gbif.org/) very simple and straightforward. However, the truth is that natural history collections are almost as far from complete digitization as they were 20 years ago. Ariño (2010)[3] estimated that no more than 3% of biological specimen data is web-accessible through GBIF, the largest source of biodiversity information. Consequently, there is neither a central database of collection holdings, nor a complete collection index available to users. The reason for this deficiency is partly the immense effort it would take to digitize the vast number of collections units involved (Vollmar et al. 2010).[4] The cost of traditional digitization workflows is vast, both in financial and human terms. Our simple calculations have shown that complete databasing of the ~30 million insect specimens housed in the entomological collection of the Natural History Museum, London, would require 23 years of continuous work from the entire departmental staff to complete (65 people). Depending on the particular collections and curatorial practices used, estimates vary from US$0.50 to several dollars per specimen to capture full label data (Heidorn 2011).[5] The cost of traditional imaging and databasing of every natural history object in all European museums was recently estimated as €73.44 per object (Poole 2010).[6] Thus, the complete digitization of all natural history collections may cost as much as €150, 000 million, and take as long as 1,500 years.
The most common solution proposed to overcome the enormous cost of digitization is prioritization based on user demand (Berents et al. 2010).[7] Currently, most digitization projects concentrate their efforts on obtaining high quality images of selected specimens accompanied by high quality data (e.g., comprehensive and expertly interpreted label information) rather than total collections coverage. Such specimen-centric digitization efforts are thus inevitably fragmented into numerous small-scale and labour-intensive projects that usually image single specimens, one at a time.
To solve the problem of cost, as well as the inherent fragmentation in collection based biodiversity informatics, new, industrial-scale approaches to digitization are clearly needed. The larger a digitization project becomes, the lower are the transaction costs and thus the lower is the cost per specimen. Such an industrial-scale process must necessarily fulfill certain standardized criteria if it is to be of use to and adopted by a wide spectrum of natural history collections:
- As much as possible of the procedure must be automated, except when physical handling of specimens is necessary.
- The approach should, whenever possible, focus on “wall-to-wall” total digitization of entire collections, because it is faster to digitize an entire collection than to select individual specimens or drawers of particular interest.
- Complicated labour-intensive procedures must be divided into a series of separate, shorter steps, each with a distinct outcome. For example, preparation of specimens for imaging should be a separate step from the imaging itself; and unique specimen identifiers can be assigned simultaneously to all specimens in a drawer rather than individually and sequentially. Such a modularised process can then be more easily crowd-sourced among the professional and volunteer communities. Properly organized crowd-sourcing projects would be able to mobilise the efforts of thousands of enthusiasts around the world (Hill et al. 2012).[8]
- Collection of metadata must be simplified and standardized. In most cases, digital representation of the specimen and minimal metadata (uID, specimen location in the collection) is sufficient for collection management purposes. Only minimal information should be collected when initially digitizing an entire collection, but in such a way that it can be amended and expanded upon later.
Here we describe a new method for “wall-to-wall” mass-digitization of natural history museum collections based on the SatScan® tray scanning system. The method allows for standardized scanning of museum collection trays of the highest image quality possible, followed by simplified (and easily expandable) collection of metadata.
Methods
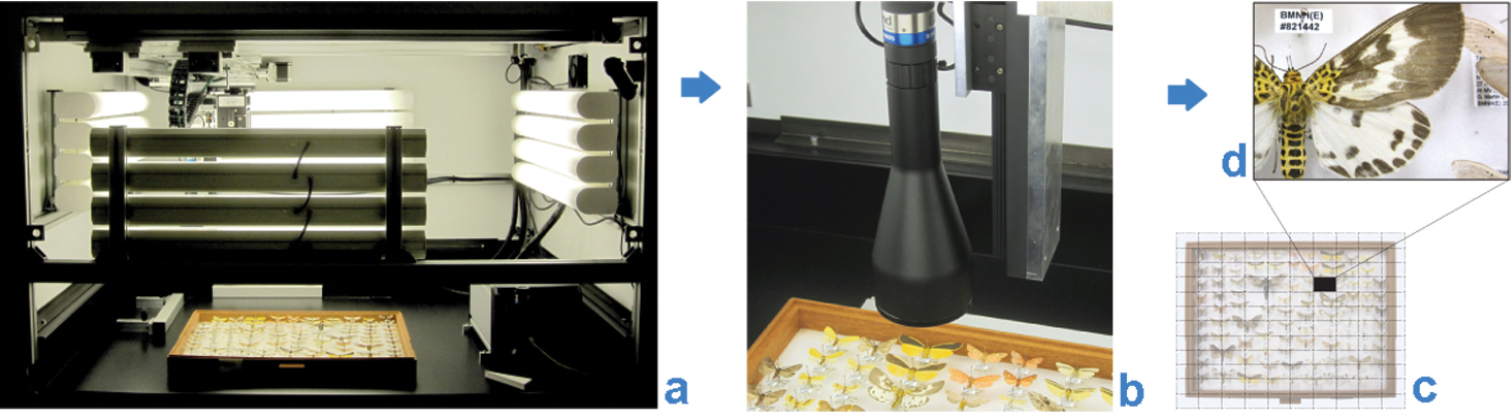
The Natural History Museum (NHM), London, has been working with SmartDrive Limited (http://www.smartdrive.co.uk/) since 2009 on the development of one of the company’s products, the SatScan® collection scanner (Fig. 1). From this collaboration, we have developed a workflow that we consider meets our needs for the industrial-scale digitization of a significant part of the NHM’s collections. The system is particularly suited to the digitization of multiple, uniformly mounted or laid out specimens, such as pinned insects and smaller geological or mineralogical objects in standardized collection drawers, horizontally-stored microscope slides and herbarium sheets.
|
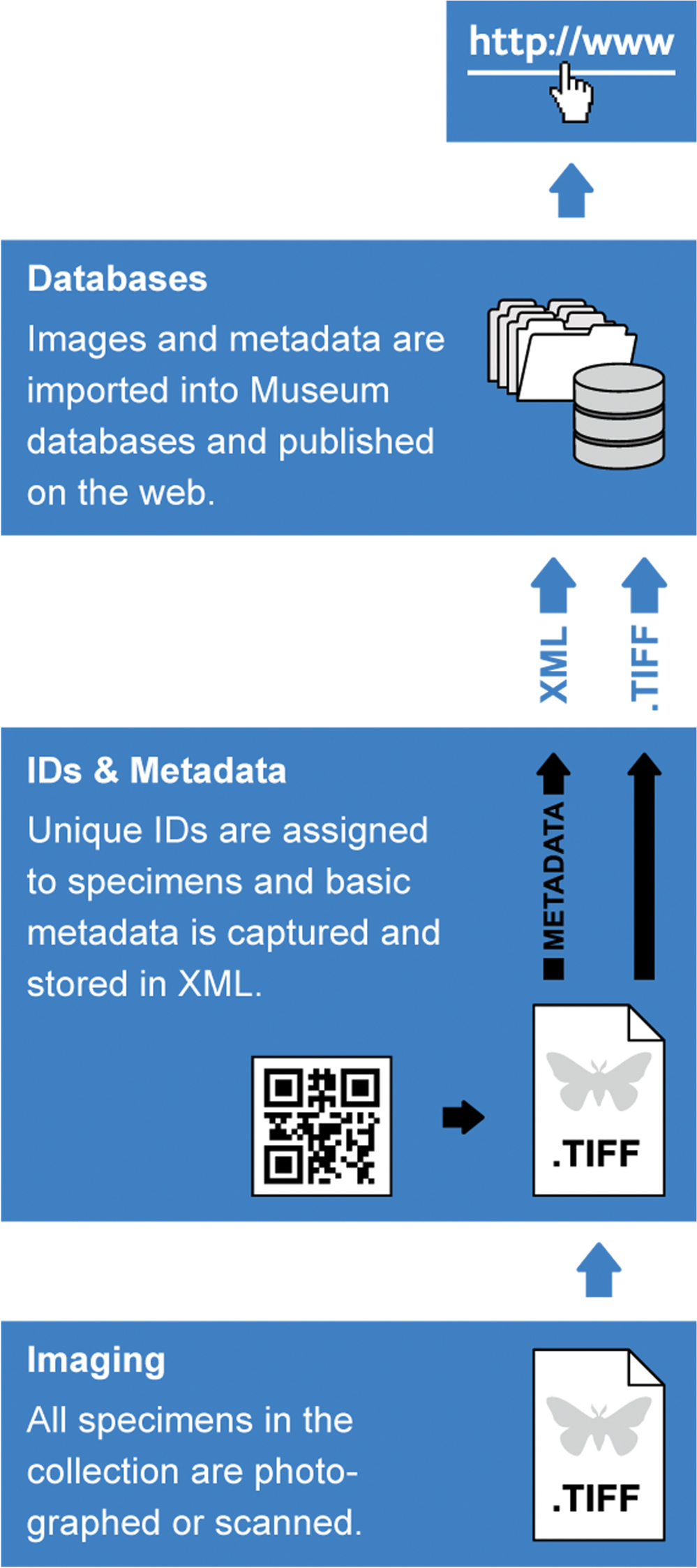
The digitization workflow envisioned for the NHM (Fig. 2) comprises three steps:
|
Imaging
The SatScan® collection scanner is capable of producing high-resolution images of entire collection drawers (see Table 1, Blagoderov et al. 2010, Mantle et al. 2012).[9][10] The specific configuration of the system has changed somewhat from that described in the report, such that now a USB CMOS UEye-SE camera (model # UI-1480SE-C-HQ, 2560×1920 resolution) is used in combination with Edmund Optics telecentric TML lenses of 0.3× (#58428) and 0.16× TML (#56675). A camera with attached lens is moved in two dimensions along precision-engineered rails positioned above the object to be imaged. A combination of hardware and software provides automated capture of high resolution images of small regions of interest, which are then assembled (“stitched”) into a larger panoramic image, generating the final image of the entire drawer. This method maximizes depth of field of the captured images and minimizes distortion and parallax artefacts. Analogous solutions for large-area imaging which have been developed independently include GigaPan (Bertone et al. 2012)[11], MicroGigaPan (Longson et al. 2010)[12] and DScan (Schmidt et al. 2012).[13]
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Metadata capture
A prototype software program, Metadata Creator, has been designed to allow fast capture of specimen data and associating these with the image of the specimen (Fig. 3). Users can mark individual specimens on the panoramic image by drawing rectangular boxes around them, selecting these areas and annotating them individually or in batches. Methods for marking the specimen, editing regions of interest and selection of multiple specimens are analogous to those used in many common graphic applications and so will be familiar, even to inexperienced users.
Specimen metadata is captured in a series of fields that are compatible with the Darwin Core 1.4.1 schema (http://rs.tdwg.org/dwc/) and which can be customized to particular user requirements. To maximize throughput, only basic metadata are collected at this stage. These will generally include a unique collection number of every specimen (see below, barcodes), collection identification (to the available curatorial level, e.g. to species/subspecies for the “Main Collection” and family/order for unsorted accessions), and, if possible, biogeographic region/country. Taxon names are looked up from an index derived from the NHM Collections Management Database. A completed project comprises a folder with an archival image of the drawer, full-resolution images of individual specimens cut-out from the drawer image, and an XML file containing annotations and links to specimen images (Appendix 1). Trials have demonstrated that 10–20 seconds per specimen is required to capture basic metadata using the Metadata Creator Software. A unique ID for the drawer is also recorded. As the NHM Collection Management System already includes a complete collections index (a brief description of the content of every drawer), no additional information is required.
|
Assigning uIDs
Every specimen is assigned a unique number under which it will be registered in the NHM Collections Management Database. It is a requirement of collections management procedures that a label bearing the specimen’s uID is attached to the specimen. To streamline this part of the process, it is subdivided into the following steps:
- A sequence of unique numbers is generated from the NHM Collections Management Database.
- Labels that include both a human-readable number and a machine-readable barcode are printed.
- The operator labels the specimens by selecting a specimen on the drawer image, pinning a label under the specimen, and scanning the barcode, thereby adding the uID into the corresponding field of Metadata Creator. Barcodes can be pinned facing up or down depending on curatorial practice; the former has the advantage of visibility on the image. In this case imaging, of course, has to take place after assigning uIDs. Images of individual specimens for which the metadata have been collected and individual numbers assigned are automatically marked on the drawer image with a grey spot, allowing easy visualization of progress.
- When all specimens have been labelled and recorded, the XML file and corresponding specimen images are imported into the NHM Collections Management Database.
We must emphasize that Metadata Creator is a prototype software application; much more development is needed for to perfect its functionality, user interface, and integration with the Museum’s information systems.
References
- ↑ Chapman, A. (2005). "Uses of Primary Species-Occurrence Data, version 1.0". Report for the Global Biodiversity Information Facility, Copenhagen. Global Biodiversity Information Facility. pp. 100. http://www.gbif.org/resource/80545.
- ↑ Baird, R. (2010). "Leveraging the fullest potential of scientific collections through digitization". Biodiversity Informatics 7 (2): 130–136. doi:10.17161/bi.v7i2.3987.
- ↑ 3.0 3.1 Ariño, A.H. (2010). "Approaches to estimating the universe of natural History collections data". Biodiversity Informatics 7 (2): 81–92. doi:10.17161/bi.v7i2.3991.
- ↑ Vollmar, A.; Macklin, J.A.; Ford, L. (2010). "Natural history specimen digitization: Challenges and concerns". Biodiversity Informatics 7 (2): 93–112. doi:10.17161/bi.v7i2.3992.
- ↑ Heidorn, P.B. (2011). "Biodiversity informatics". Bulletin of the American Society for Information Science and Technology 37 (6): 38–44. doi:10.1002/bult.2011.1720370612.
- ↑ Poole, N. (November 2010). "The Cost of Digitising Europe's Cultural Heritage". Collections Trust. pp. 82. http://www.collectionstrust.org.uk/item/739-the-cost-of-digitising-europe-s-cultural-heritage.
- ↑ Berents, P.; Hamer, M.; Chavan, V. (2010). "Towards demand driven publishing: Approches to the prioritisation of digitisation of natural history collections data". Biodiversity Informatics 7 (2): 113–119. doi:10.17161/bi.v7i2.3990.
- ↑ Hill, A.; Guralnick, R.; Smith, A. (2012). "The notes from nature tool for unlocking biodiversity records from museum records through citizen science". ZooKeys 209: 219–233. doi:10.3897/zookeys.209.3472.
- ↑ Blagoderov, V.; Kitching, I.; Simonsen, T.; Smith, V. (2010). "Report on trial of SatScan tray scanner system by SmartDrive Ltd.". Nature Precedings: 7. http://precedings.nature.com/documents/4486/version/1.
- ↑ Mantle, B.L.; La Salle, J.; Fisher, N. (2012). "Whole-drawer imaging for digital management and curation of a large entomological collection". ZooKeys 209: 147–163. doi:10.3897/zookeys.209.3169.
- ↑ Bertone, M.A.; Blinn, R.L.; Stanfield, T.M. et al. (2012). "Results and insights from the NCSU Insect Museum GigaPan project". ZooKeys 209: 115–132. doi:10.3897/zookeys.209.3083.
- ↑ Longson, J.; Cooper, G.; Gibson, R. et al. (2010). "Adapting Traditional Macro and Micro Photography for Scientific Gigapixel Imaging". Proceedings of the Fine International Conference on Gigapixel Imaging for Science. http://repository.cmu.edu/gigapixel/1/.
- ↑ Schmidt, S.; Balke, M.; Lafogler, S. (2012). "DScan – a high-performance digital scanning system for entomological collections". ZooKeys 209: 183–191. doi:10.3897/zookeys.209.3115.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. Additionally, a missing reference (Vollmar et al. 2010) was added.