Journal:OpenChrom: A cross-platform open source software for the mass spectrometric analysis of chromatographic data
| Full article title | OpenChrom: A cross-platform open source software for the mass spectrometric analysis of chromatographic data |
|---|---|
| Journal | BMC Bioinformatics |
| Author(s) | Wenig, Philip; Odermatt, Juergen |
| Author affiliation(s) | University of Hamburg |
| Primary contact | Email: philip.wenig@gmx.net |
| Year published | 2010 |
| Volume and issue | 11 |
| Page(s) | 405 |
| DOI | 10.1186/1471-2105-11-405 |
| ISSN | 1471-2105 |
| Distribution license | Creative Commons Attribution 2.0 Generic |
| Website | http://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-11-405 |
| Download | http://bmcbioinformatics.biomedcentral.com/track/pdf/10.1186/1471-2105-11-405 (PDF) |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
Background
Today, data evaluation has become a bottleneck in chromatographic science. Analytical instruments equipped with automated samplers yield large amounts of measurement data, which needs to be verified and analyzed. Since nearly every GC/MS instrument vendor offers its own data format and software tools, the consequences are problems with data exchange and a lack of comparability between the analytical results. To challenge this situation a number of either commercial or non-profit software applications have been developed. These applications provide functionalities to import and analyze several data formats but have shortcomings in terms of the transparency of the implemented analytical algorithms and/or are restricted to a specific computer platform.
Results
This work describes a native approach to handle chromatographic data files. The approach can be extended in its functionality such as facilities to detect baselines, to detect, integrate and identify peaks and to compare mass spectra, as well as the ability to internationalize the application. Additionally, filters can be applied on the chromatographic data to enhance its quality, for example to remove background and noise. Extended operations like do, undo and redo are supported.
Conclusions
OpenChrom is a chromatography software application to edit and analyze mass spectrometric chromatographic data. It is extensible in many different ways, depending on the demands of the users or the analytical procedures and algorithms. It offers a customizable graphical user interface. The software is independent of the operating system, due to the fact that the Rich Client Platform is written in Java. OpenChrom is released under the Eclipse Public License 1.0 (EPL). There are no license constraints regarding extensions. They can be published using open source as well as proprietary licenses. OpenChrom is available free of charge at http://www.openchrom.net.
Background
Software has become an integral part of analysis techniques. Especially in the area of gas chromatography/mass spectrometry, automatic samplers enable high throughput analyses. Software assists handling large amounts of data generated by automated and fast operating analytical instruments. Modern computer systems are inexpensive, powerful and allow analysis techniques that could not have been applied in the past. Deconvolution, a chromatographic quality enhancing technique, demonstrates for instance that increasing processor power makes new analysis techniques applicable. The technique of deconvolution has been described by Biller and Biemann[1][2], Dromey et al.[3], Colby[4], Hindmarch et al.[5], Halket et al.[6], Kong et al.[7], Taylor et al.[8], Pool et al.[9][10] and Davies[11] in various ways. Stein[12] published an enhanced deconvolution algorithm that has been implemented in the software AMDIS (Automated Mass Spectral Deconvolution and Identification System).[13] AMDIS is available free of charge from the National Institute of Standards and Technology (NIST). Windig et al.[14][15] described another approach to enhance chromatographic quality by a deconvolution method called CODA (Component Detection Algorithm). The commercially available software ACD/MS Manager[16] offers an implementation of this approach.
Increasing computational power enables new applications, but there is still a lack of interoperability. Instrument vendors, such as Agilent Technologies, Shimadzu, Thermo Fisher Scientific and Waters Corporation have created their own software and data format. Usually, the mass spectral data formats are binary and can only be accessed by the instrument vendors' proprietary software. Some commercial tools exist to convert the mass spectral data files into other formats, such as MASS Transit from PALISADE Corporation.[17] To avoid these limitations, some efforts have been made to design and implement interoperable data formats and software libraries as for example NetCDF[18] or mzXML.[19][20] But even if it is possible to convert the data files to other formats, there are drawbacks in data processing as each software implements specific functions, has its own graphical user interface and is in most cases commercially available only, as for example the applicable software of ChemStation, Xcalibur or MassLynx. Hence, the users are forced to become familiar with different software systems, user interfaces and methods. Moreover, the software tools primarily target only specific operating systems, such as Microsoft Windows and Mac OSX. The number of software applications that are independent of the operating system and can also be run under Unix or Linux is limited. Linux systems are open source, available at no cost and their usage increases in scientific research (see Scientific Linux[21]), as well as in the public sector.[22][23] Software applications, such as AMDIS, have been published to be used free of charge, but their source code is not disposable. Thus, it is not possible to evaluate the algorithms implemented in the software. Especially in the case of scientific research, it is not possible to figure them out and to extend them. Even if algorithms are described in published papers[2][4][9][12][24], it is often impossible to validate them manually due to the complexity of chromatographic data. Other applications like ChemStation, Xcalibur, and ACD/MS Manager are proprietary and closed source. They are only commercially available. There is no means of revealing the correctness of their utilized algorithms. Efforts have been made to solve the problems of missing interoperability and restricted access to source codes and algorithms.[25] Bioclipse is a sophisticated project that is open source and is focused with its algorithms on metabolism analysis and gene sequencing. Its techniques are state-of-the-art. Some other projects are mMass[26], COMSPARI[27] and fityk[28], but they do have some restrictions regarding their interoperability and extensibility. BioSunMS[29] is a tool to read TOF (Time of Flight) mass spectral data files, but it is not able to read instrument vendors' native data files. The Chemistry Development Kit (CDK)[30] implements convenient features to edit chemical data and structures, but it has no appropriate user interface. The open source tool OpenMS[31] aims to edit mass spectrometric data, but it is not completely platform independent, as it is written in C++ programming language.
Projects like Bioclipse, Sashimi[32] or TPP (Trans-Proteomic Pipeline)[33] are focused on the evaluation of metabolism products and gene sequencing and make extensive use of accurate mass resolution techniques. But there is still a lack of software systems that are capable to enhance nominal mass spectral data files, that are flexible, extensible and that offer an easy to use graphical user interface. According to the authors' knowledge, no application offers functions to import vendor systems chromatographic data files and has the ability to edit and analyze chromatograms in the way ChemStation and AMDIS do. No application combines the flexibility in analyses, is easily extensible, open source, platform independent and has a configurable graphical user interface.
Implementation
Architecture
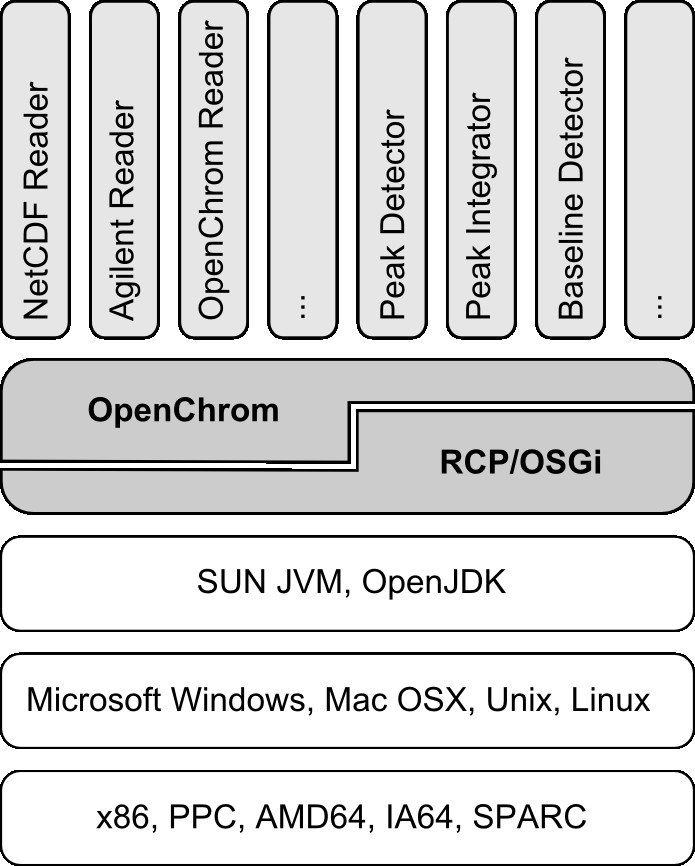
OpenChrom is an open source software that aims to solve the aforementioned constraints getting rid of several restrictions. It is based on the Eclipse Rich Client Platform (RCP)[34], which is an OSGi (Open Service Gateway Initiative) based application environment that allows to build modular and flexible software systems. With the OSGi platform it is possible to extend the functionality of an application by dividing its components into different bundles. It is written in Java which is an interpreted language that depends on the Java Virtual Machine (JVM) and allows the execution of the software on several operating systems (Microsoft Windows, Mac OSX, Unix, Linux) and processor platforms (x86, PPC, AMD64, IA64, SPARC). It utilizes SWT (Standard Widget Toolkit) to render its graphical user interface by using the native resources of the underlying operating system. The Rich Client Platform is state-of-the-art in today's software development. The platform is open to be extended afterwards due to the chosen concepts. It means that the platform doesn't need to be full-fledged at the beginning. Further methods and implementations can be developed separately. Nonetheless, still some effort is necessary to design a platform that covers all needs of a software application to edit, evaluate and modify chromatographic data. In contrast to Bioclipse, Sashimi or TPP, OpenChrom has a slightly different scope, as it is focused primarily on nominal mass resolution data. Mass spectrometers for nominal mass resolution are inexpensive, as for example quadrupole or ion trap instruments. But the data acquisition limits the range of possible applications. Software has the potential to enhance the quality of the recorded data, in contrary to the given limitations. Hence, the Rich Client Platform and the Java programming language were chosen, as they offer an excellent support for a highly extensible and abstract base framework. The OSGi based Rich Client Platform Equinox supports the definition of extension points. The use of different class paths makes it possible to execute code from separated bundles (Figure 1). New functionality, e.g. to export a given chromatogram to a PDF file, can be implemented in a separate bundle making use of the extension point mechanism to import and export chromatographic data.
|
Tools in different areas have been implemented based on the Rich Client Platform, such as the Eclipse IDE (Integrated Development Environment), Lotus Notes, Bioclipse, BioSunMS, XMind, Apache Directory Studio and several more. It is part of the OpenChrom architecture to define useful extension points and to build a suitable object model.
Object model
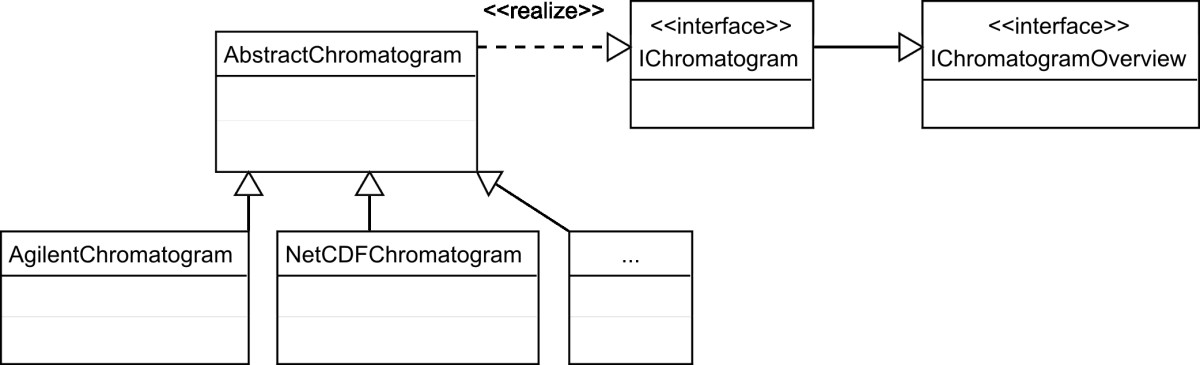
OpenChrom provides a designed object model to define chromatograms, scans, mass spectra, peaks and baselines. It is important to abstract the base model, as it reduces dependencies in code and allows for the implemention of further extensions. Therefore, the decision was to support an enhanced chromatogram, mass spectrum and peak model, written in Java. There is no preliminary compilation necessary on different operating systems. Further on, it is possible to cover special needs regarding the import of instrument vendors' binary chromatographic files. An excerpt of the OpenChrom object model is shown in a simplified UML (Unified Modeling Language) diagram (Figure 2). Java, as an object orientated language, supports the use of the four base strategies in object orientation: abstraction, encapsulation, polymorphic behavior and inheritance.[35] OpenChrom makes extensive use of the object orientated concept. The interface "IChromatogram" and the abstract class "AbstractChromatogram" define and implement methods, which are common for all types of chromatograms, independent of the instrument vendors' data format. Therefore, it is not necessary to implement them iterative in each vendor specific chromatogram class. The base framework and extension points, like peak detectors and integrators, are working still with instances of the type "IChromatogram", instead of taking for example the differences of an Agilent and a NetCDF chromatogram into account. The object model for mass spectra and mass fragments, peaks and baselines is implemented in a similar way.
|
Extension points
The OpenChrom framework offers several bundles (Table 1). The most important one defines methods to implement specialized bundles that handle the import of chromatographic mass spectral data. It is possible to supply a bundle that is able to read binary chromatogram files, given by a specific instrument vendor. The bundle takes care of how to read a given file or directory. Furthermore, the framework offers extension points to detect and integrate peaks. The peak detection and integration have been separated, to make it possible to detect peaks with several peak detector methods and to integrate them with a specified integrator. This results in a more complex but also more flexible system. There is another extension point that allows to define bundles that are capable of detecting a baseline in the chromatogram model. Another flexible extension point was introduced, called filters. Bundles can extend the filter extension point to achieve a quality enhancement of the chromatographic data. They work comparable to filters in image processing software. One filter extension can for instance offer a set of methods to eliminate background signals from the chromatogram. Another filter can implement a routine to mean normalize the chromatogram. The filters offer editing steps, which are especially useful before peak detection and integration routines.
| ||||||||||||||||||||||||||||||
Graphical user interface
The Rich Client Platform offers a wide support to present an appropriate graphical user interface. Concepts detailing this include editors, views, perspectives, wizards, menus, cheat sheets, settings and help pages. OpenChrom makes extensive use of the available concepts. The editor shows the graphical representation of a chromatogram and several options, as for example a page to select or exclude distinct mass fragments. It also supports functions to save, edit and analyze chromatograms. The views are used to show different aspects of the chromatographic model. It is possible to show peaks in different kind of views. One view could show a peak including the background of the chromatogram. Another could show the peak with its increasing and decreasing tangents and its width at 50% height. A flexible mechanism was introduced to inform all views if the chromatogram selection has been changed. The update functionality is also realized by an extension point. Views and editors are composed in a task specific way using perspectives.
References
- ↑ Biller, J.E.; Herlihy, W.C.; Biemann, K. (1977). "Identification of the components of complex mixtures by GC-MS". Abstracts Of Papers Of The American Chemical Society 173 (MAR20): 23–23. http://pubs.acs.org/doi/abs/10.1021/bk-1977-0054.ch002.
- ↑ 2.0 2.1 Biller, J.E.; Biemann, K. (1974). "Reconstructed Mass Spectra, A Novel Approach for the Utilization of Gas Chromatograph—Mass Spectrometer Data". Analytical Letters 7 (7): 515–528. doi:10.1080/00032717408058783.
- ↑ Dromey, R.G.; Stefik, M.J.; Rindfleisch, T.C.; Duffield, A.M. (1976). "Extraction of mass spectra free of background and neighboring component contributions from gas chromatography/mass spectrometry data". Analytical Chemistry 48 (9): 1368–1375. doi:10.1021/ac50003a027.
- ↑ 4.0 4.1 Colby, B.N. (1992). "Spectral deconvolution for overlapping GC/MS components". Journal of the American Society for Mass Spectrometry 3 (5): 558–562. doi:10.1016/1044-0305(92)85033-G. PMID 24234499.
- ↑ Hindmarch, P.; Demir, C.; Brereton, R.G. (1996). "Deconvolution and spectral clean-up of two-component mixtures by factor analysis of gas chromatographic–mass spectrometric data". Analyst 121 (8): 993-1001. doi:10.1039/AN9962100993.
- ↑ Halket, J.M.; Przyborowska, A.; Stein, S.E. et al. (1999). "Deconvolution gas chromatography/mass spectrometry of urinary organic acids – potential for pattern recognition and automated identification of metabolic disorders". Rapid Communications In Mass Spectrometry 13 (4): 279–284. doi:10.1002/(SICI)1097-0231(19990228)13:4<279::AID-RCM478>3.0.CO;2-I. PMID 10097403.
- ↑ Kong, H.W.; Ye, F.; Lu, X.; Guo, L.; Tian, J.; Xu, G.W. (2005). "Deconvolution of overlapped peaks based on the exponentially modified Gaussian model in comprehensive two-dimensional gas chromatography". Journal of Chromatography A 1086 (1–2): 160–164. doi:10.1016/j.chroma.2005.05.103. PMID 16130668.
- ↑ Taylor, J.; Goodacre, R.; Wade, W.G. (1998). "The deconvolution of pyrolysis mass spectra using genetic programming: Application to the identification of some Eubacterium species". FEMS Microbiology Letters 160 (2): 237–246. doi:10.1111/j.1574-6968.1998.tb12917.x. PMID 9532743.
- ↑ 9.0 9.1 Pool, W.G.; deLeeuw, J.W.; vandeGraaf, B. (1996). "Backfolding applied to differential gas chromatography/mass spectrometry as a mathematical enhancement of chromatographic resolution". Journal Of Mass Spectrometry 31 (5): 509–516. doi:10.1002/(SICI)1096-9888(199605)31:5<509::AID-JMS323>3.0.CO;2-B.
- ↑ Pool, W.G.; deLeeuw, J.W.; vandeGraaf, B. (1997). "Automated extraction of pure mass spectra from gas chromatographic/mass spectrometric data". Journal Of Mass Spectrometry 32 (4): 438–443. doi:10.1002/(SICI)1096-9888(199704)32:4<438::AID-JMS499>3.0.CO;2-N.
- ↑ Davies, A. (1998). "The new Automated Mass Spectrometry Deconvolution and Identification System (AMDIS)". Spectrometry Europe 10 (3): 22–26.
- ↑ 12.0 12.1 Stein, S.E. (1999). "An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data". Journal Of the American Society for Mass Spectrometry 10 (8): 770–781. doi:10.1016/S1044-0305(99)00047-1.
- ↑ "AMDIS". The National Institute of Standards and Technology. http://chemdata.nist.gov/dokuwiki/doku.php?id=chemdata:amdis.
- ↑ Windig, W.; Smith, W.F. (2007). "Chemometric analysis of complex hyphenated data: Improvements of the component detection algorithm". Journal of Chromatography A 1158 (1–2): 251–257. doi:10.1016/j.chroma.2007.03.081. PMID 17418223.
- ↑ Windig, W.; Phalp, J.M.; Payne, A.W. (1996). "A noise and background reduction method for component detection in liquid chromatography/mass spectrometry". Analytical Chemistry 68 (20): 3602–3606. doi:10.1021/ac960435y.
- ↑ "ACD/Labs". Advanced Chemistry Development, Inc. http://www.acdlabs.com/.
- ↑ "PALISADE". Palisade Corporation. http://www.palisade.com/.
- ↑ "Network Common Data Form (NetCDF)". University Corporation for Atmospheric Research. http://www.unidata.ucar.edu/software/netcdf/.
- ↑ Pedrioli, P.G.A.; Eng, J.K.; Hubley, R. (2004). "A common open representation of mass spectrometry data and its application to proteomics research". Nature Biotechnology 22 (11): 1459–1466. doi:10.1038/nbt1031. PMID 15529173.
- ↑ Falkner, J.A.; Falkner, J.W.; Andrews, P.C. (2007). "ProteomeCommons.org IO Framework: Reading and writing multiple proteomics data formats". Bioinformatics 23 (2): 262–263. doi:10.1093/bioinformatics/btl573. PMID 17121776.
- ↑ "Scientific Linux". Wikipedia. Wikimedia Foundation, Inc. https://en.wikipedia.org/wiki/Scientific_Linux.
- ↑ "Wienux". Wikipedia. Wikimedia Foundation, Inc. https://en.wikipedia.org/wiki/Wienux.
- ↑ "Das Projekt LiMux". Portal München Betriebs-GmbH & Co. KG. http://www.muenchen.de/rathaus/Stadtverwaltung/Direktorium/LiMux.html.
- ↑ Alfassi, Z.B. (2004). "On the normalization of a mass spectrum for comparison of two spectra". Journal of the American Society for Mass Spectrometry 15 (3): 385-387. doi:10.1016/j.jasms.2003.11.008. PMID 14998540.
- ↑ Spjuth, O.; Helmus, T.; Willighagen, E.L. et al. (2007). "Bioclipse: An open source workbench for chemo- and bioinformatics". BMC Bioinformatics 8: 59. doi:10.1186/1471-2105-8-59. PMC PMC1808478. PMID 17316423. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1808478.
- ↑ "mMass - Open Source Mass Spectrometry Tool". Martin Strohalm. Archived from the original on 27 August 2009. https://web.archive.org/web/20090827071924/http://mmass.biographics.cz/.
- ↑ "The COMSPARI Homepage". J. Katz and J. Hau. http://www.biomechanic.org/comspari/.
- ↑ "Fityk home". Institute of High Pressure Physics of the Polish Academy of Sciences. Archived from the original on 04 March 2010. https://web.archive.org/web/20100304192315/http://www.unipress.waw.pl/fityk.
- ↑ Cao, Y.; Wang, N.; Ying, X.M. et al. (2009). "BioSunMS: A plug-in-based software for the management of patients information and the analysis of peptide profiles from mass spectrometry". BMC Medical Informatics and Decision Making 9: 13. doi:10.1186/1472-6947-9-13. PMC PMC1808478. PMID 17316423. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1808478.
- ↑ Steinbeck, C.; Han, Y.Q.; Kuhn, S. et al. (2003). "The Chemistry Development Kit (CDK): An open-source Java library for Chemo- and Bioinformatics". Journal of Chemical Information and Computer Sciences 43 (2): 493–500. PMID 12653513.
- ↑ Sturm, M.; Bertsch, A.; Gropl, C. et al. (2008). "OpenMS – An open-source software framework for mass spectrometry". BMC Bioinformatics 9: 163. doi:10.1186/1471-2105-9-163. PMC PMC2311306. PMID 18366760. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2311306.
- ↑ "Sashimi". SourceForge. http://sourceforge.net/projects/sashimi/.
- ↑ "Seattle Proteome Center (SPC) - Proteomics Tools". Institute for System Biology. http://tools.proteomecenter.org/.
- ↑ "Rich Client Platform". The Eclipse Foundation. http://wiki.eclipse.org/Rich_Client_Platform.
- ↑ Horstmann, C.S.; Cornell, G. (2001). Core Java 2: Fundamentals. Upper Saddle River, NJ: Prentice Hall Professional. pp. 806. ISBN 9780130894687. https://books.google.com/books?id=W6bomXWB-TYC.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. In the "Conclusion" section of the abstract, "software" was changed to "chromatography software" to encourage internal linking to the CDMS entry on the wiki. Two blank rows were removed from Table 1.