Journal:Popularity and performance of bioinformatics software: The case of gene set analysis
| Full article title | Popularity and performance of bioinformatics software: The case of gene set analysis |
|---|---|
| Journal | BMC Bioinformatics |
| Author(s) | Xie, Chengshu; Jauhari, Shaurya; Mora, Antonio |
| Author affiliation(s) | Guangzhou Medical University, Guangzhou Institutes of Biomedicine and Health |
| Primary contact | Online form |
| Year published | 2017 |
| Volume and issue | 22 |
| Article # | 191 |
| DOI | 10.1186/s12859-021-04124-5 |
| ISSN | 1471-2105 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-021-04124-5 |
| Download | https://bmcbioinformatics.biomedcentral.com/track/pdf/10.1186/s12859-021-04124-5.pdf (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
Background: Gene set analysis (GSA) is arguably the method of choice for the functional interpretation of omics results. This work explores the popularity and the performance of all the GSA methodologies and software published during the 20 years since its inception. "Popularity" is estimated according to each paper's citation counts, while "performance" is based on a comprehensive evaluation of the validation strategies used by papers in the field, as well as the consolidated results from the existing benchmark studies.
Results: Regarding popularity, data is collected into an online open database ("GSARefDB") which allows browsing bibliographic and method-descriptive information from 503 GSA paper references; regarding performance, we introduce a repository of Jupyter Notebook workflows and shiny apps for automated benchmarking of GSA methods (“GSA-BenchmarKING”). After comparing popularity versus performance, results show discrepancies between the most popular and the best performing GSA methods.
Conclusions: The above-mentioned results call our attention towards the nature of the tool selection procedures followed by researchers and raise doubts regarding the quality of the functional interpretation of biological datasets in current biomedical studies. Suggestions for the future of the functional interpretation field are made, including strategies for education and discussion of GSA tools, better validation and benchmarking practices, reproducibility, and functional re-analysis of previously reported data.
Keywords: bioinformatics, pathway analysis, gene set analysis, benchmark, GSEA
Background
Bioinformatics method and software selection is an important problem in biomedical research, due to the possible consequences of choosing the wrong methods among the existing myriad of computational methods and software available. Errors in software selection may include the use of outdated or suboptimal methods (or reference databases) or misunderstanding the parameters and assumptions behind the chosen methods. Such errors may affect the conclusions of the entire research project and nullify the efforts made on the rest of the experimental and computational pipeline.[1]
This paper discusses two main factors that motivate researchers to make method or software choices, that is, the popularity (defined as the perceived frequency of use of a tool among members of the community) and the performance (defined as a quantitative quality indicator measured and compared to alternative tools). This study is focused on the field of “gene set analysis” (GSA), where the popularity and performance of bioinformatics software show discrepancies, and therefore the question appears whether biomedical sciences have been using the best available GSA methods or not.
GSA is arguably the most common procedure for functional interpretation of omics data, and, for the purposes of this paper, we define it as the comparison of a query gene set (a list or a rank of differentially expressed genes, for example) to a reference database, using a particular statistical method, in order to interpret it as a rank of significant pathways, functionally related gene sets, or ontology terms. Such definition includes the categories that have been traditionally called "gene set analysis," "pathway analysis," "ontology analysis," and "enrichment analysis." All GSA methods have a common goal, which is the interpretation of biomolecular data in terms of annotated gene sets, while they differ depending on characteristics of the computational approach (for more details, see the "Methods" section, as well as Fig. 1 of Mora[2]).
GSA has arrived to 20 years of existence since the original paper of Tavazoie et al.[3], and many statistical methods and software tools have been developed during this time. A popular review paper listed 68 GSA tools[4], while a second review reported an additional 33 tools[5], and a third paper 22 tools.[6] We have built the most comprehensive list of references to date (503 papers), and we have quantified each paper’s influence according to their current number of citations (see Additional file 1 and Mora's GSARefDB[7]). The most common GSA methods include Over-Representation Analysis (ORA), such as that found with DAVID[8]; Functional Class Scoring (FCS), such as that found with GSEA[9]; and Pathway-Topology-based (PT) methods, such as that found with SPIA.[10] All been extensively reviewed. In order to know more about them, the reader may consult any of the 62 published reviews documented in Additional file 1. We have also recently reviewed other types of GSA methods.[2]
The first part of the analysis is a study on GSA method and software popularity based on a comprehensive database of 503 papers for all methods, tools, platforms, reviews, and benchmarks of the GSA field, collectively cited 134,222 times between 1999 and 2019, including their popularity indicators and other relevant characteristics. The second part is a study on performance based on the validation procedures reported in the papers, introducing new methods together with all the existing independent benchmark studies in the above-mentioned database. Instead of recommending one single GSA method, we focus on discussing better benchmarking practices and generating benchmarking tools that follow such practices. Together, both parts of the study allow us to compare the popularity and performance of GSA tools but also to explore possible explanations for the popularity phenomenon and the problems that limit the execution and adoption of independent performance studies. In the end, some practices are suggested to guarantee that bioinformatics software selection is guided by the most appropriate metrics.
Results
Popularity
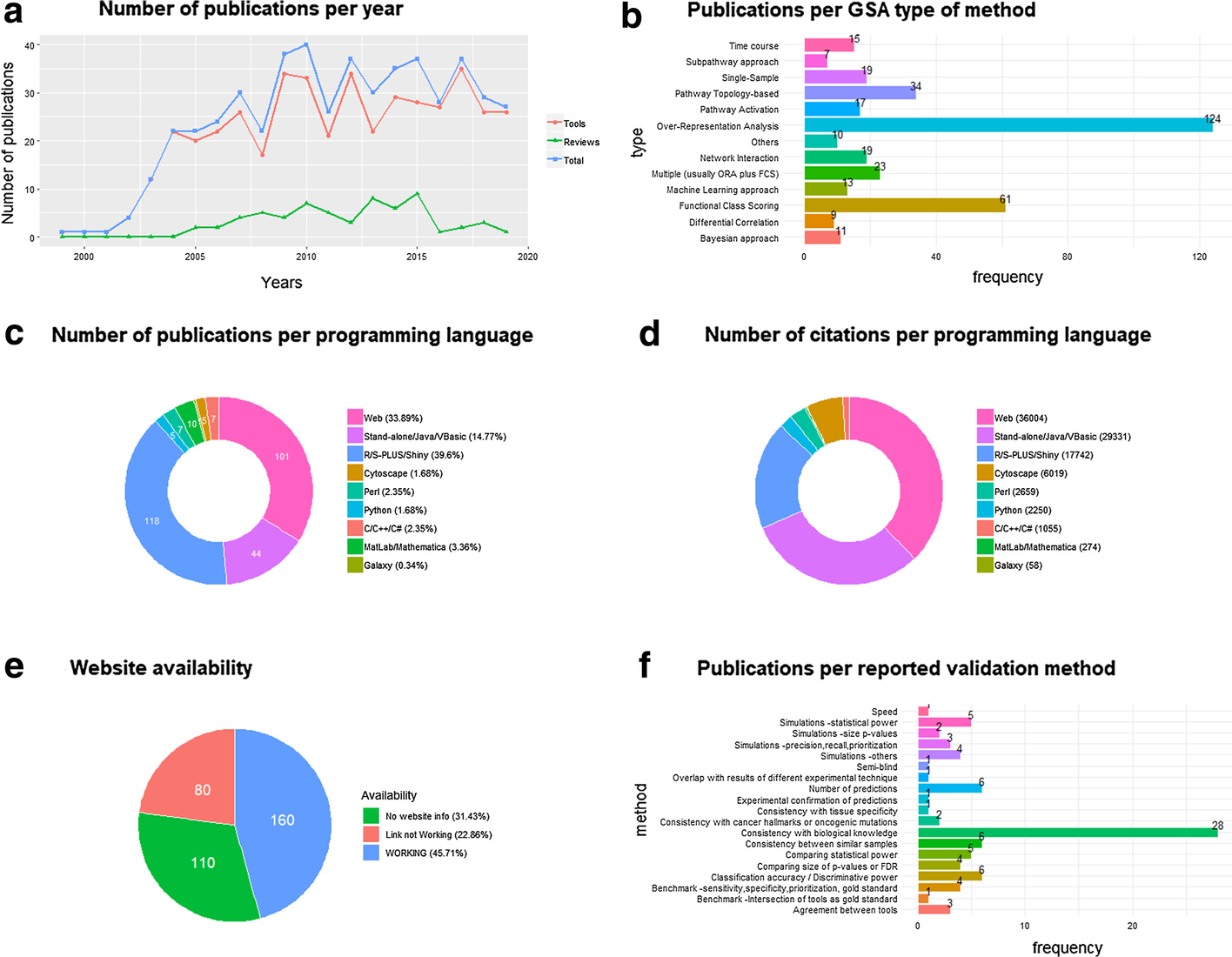
The number of paper citations has been used as a simple (yet imperfect) measure of a GSA method’s popularity. We collected 350 references to GSA papers of methods, software or platforms (Additional file 1: Tab 1), as well as 91 references to GSA papers for non-mRNA omics tools (Additional file 1: Tabs 3, 4), and 62 GSA reviews or benchmark studies (Additional file 1: Tabs 2, 3, 4), which have been organized into a manually curated open database (GSARefDB). GSARefDB can be either downloaded or accessed through a web interface. Figure 1 summarizes some relevant information from the database (GSARefDB v.1.0).
|
The citation count shows that the most influential GSA method in history is Gene Set Enrichment Analysis (GSEA), published in 2003, with more than twice as many citations as its follower, the ORA platform called DAVID (17,877 versus 7,500 citations). In general, the database shows that the field contains a few extremely popular papers and many papers with low popularity. Figure 1b shows that the list of tools is mainly composed of ORA and FCS methods, while the newer and lesser known PT and Network Interaction (NI) methods are less common (and generally found at the bottom of the popularity rank).
It could be hypothesized that the popularity of a GSA tool does not always depend on being the best for that particular project, and it could be related to variables such as its user-friendliness instead. The database allows us to compute citations-per-programming-language, which we use as an approximation to friendliness. Figure 1c shows that the majority of the GSA papers correspond to R tools, but, in spite of that, Fig. 1d shows that most of the citations correspond to web platforms followed by stand-alone applications, which are friendlier to users. Worth mentioning, the last column of the database shows that there are a large number of tools that are not maintained anymore, with broken web links to tools or databases, which makes their evaluation impossible. This phenomenon is a common bioinformatics problem that has also been reported elsewhere.[11] Figure 1e shows that one-third of the reported links in GSA papers are now broken links.
Besides ranking papers according to their all-time popularity, a current-popularity rank was also built (Additional file 1: Tab 5). To accomplish that, a version of the database generated in May 2018 was compared to a version of the database generated in April 2019. The "current popularity" rank revealed the same trends as the "overall" rank. GSEA is still, by far, the most popular method. The other tools that are being currently cited are clusterProfiler[12], Enrichr[13], GOseq[14], DAVID[8], and ClueGO[15], followed by GOrilla, KOBAS, BiNGO, ToppGene, GSVA, WEGO, agriGO, and WebGestalt. ORA and FCS methods are still the most popular ones, with 3534 combined citations for all ORA methods and 2185 citations for all FCS methods, while PT and NI methods have 111 and 50 combined citations respectively. In contrast, single-sample methods have 278 combined citations, while time-course methods have 67. Regarding reviews, an extremely popular paper from 2009[4] is still currently the most popular, even though it doesn’t take into account the achievements of the last 10 years.
Performance
The subject of [Software verification and validation|validation]] of bioinformatics software deserves more attention.[16] A review of the scientific validation approaches followed by the top 153 GSA tool papers in the database (Additional file 1: Tab 6) found multiple validation strategies that were classified into 19 categories. 61 out of the 153 papers include a validation procedure, and the most commonly found validation strategy is “Consistency with biological knowledge,” defined as the fact that our method’s results explain the knowledge in the field better than the rival methods (which is commonly accomplished through a literature search). Other common strategies (though less common) are the comparisons of the number of hits, classification accuracy, and consistency of results between similar samples. Important strategies, such as comparing statistical power, benchmark studies, and simulations, are less used. The least used strategies include experimental confirmation of predictions and semi-blind procedures where a person collects samples and another person applies the tool to guess tissue or condition. Our results have been summarized in Fig. 1f (above) and Additional file 1: Tab 6. We can see that the frequency of use of the above-mentioned validation strategies is inversely proportional to their reliability. For example, commonly used strategies such as “consistency with biological knowledge” can be subjective, and comparing the number of hits of our method with other methods on a Venn diagram[17] is a measure of agreement, not truth. On the other hand, the least used strategies, such as experimental confirmation or benchmark and simulation studies, are the better alternatives.
The next step of our performance study was a review of all the independent benchmark and simulation studies existing in the GSA field, whose references are collected in Additional file 1: Tab 2. Table 1 summarizes 10 benchmark studies of GSA methods, with different sizes, scopes, and method recommendations. A detailed description and discussion of each of the benchmarking studies can be found in Additional file 2. The sizes of all benchmarking studies are small when compared to the amount of existing methods that we have mentioned before, while their lists of best performing methods show little overlap. Only a few methods are mentioned as best performers in more than one study, including ORA methods (such as hypergeometric)[3], FCS methods (such as PADOG)[18], SS methods (such as PLAGE)[19], and PT methods (such as SPIA/ROntoTools).[20]
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
References
- ↑ Dixson, L.; Walter, H.; Schneider, M. et al. (2014). "Retraction for Dixson et al., Identification of gene ontologies linked to prefrontal-hippocampal functional coupling in the human brain". Proceedings of the National Academy of Sciences of the United States of America 111 (37): 13582. doi:10.1073/pnas.1414905111. PMC PMC4169929. PMID 25197092. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4169929.
- ↑ 2.0 2.1 Mora, A. (2020). "Gene set analysis methods for the functional interpretation of non-mRNA data-Genomic range and ncRNA data". Briefings in Bioinformatics 21 (5): 1495-1508. doi:10.1093/bib/bbz090. PMID 31612220.
- ↑ 3.0 3.1 Tavazoie, S.; Hughes, J.D.; Campbell, M.J. et al. (1999). "Systematic determination of genetic network architecture". Nature Genetics 22 (3): 281–5. doi:10.1038/10343. PMID 10391217.
- ↑ 4.0 4.1 Huang, D.W.; Sherman, B.T.; Lempicki, R.A. (2009). "Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists". Nucleic Acids Research 37 (1): 1-13. doi:10.1093/nar/gkn923. PMC PMC2615629. PMID 19033363. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2615629.
- ↑ Khatri, P.; Sirota, M.; Butte, A.J. (2012). "Ten years of pathway analysis: Current approaches and outstanding challenges". PLoS Computational Biology 8 (2): e1002375. doi:10.1371/journal.pcbi.1002375. PMC PMC3285573. PMID 22383865. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3285573.
- ↑ Mitrea, C.; Taghavi, Z.; Bokanizad, B. et al. (2013). "Methods and approaches in the topology-based analysis of biological pathways". Frontiers in Physiology 4: 278. doi:10.3389/fphys.2013.00278. PMC PMC3794382. PMID 24133454. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3794382.
- ↑ Mora, A. (26 March 2021). "GSARefDB". GSA Central. https://gsa-central.github.io/gsarefdb.html.
- ↑ 8.0 8.1 Huang, D.W.; Sherman, B.T.; Tan, Q. et al. (2007). "DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists". Nucleic Acids Research 35 (Web Server Issue): W169-75. doi:10.1093/nar/gkm415. PMC PMC1933169. PMID 17576678. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1933169.
- ↑ Subramanian, A.; Tamayo, P.; Mootha, V.K. et al. (2005). "Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles". Proceedings of the National Academy of Sciences of the United States of America 102 (43): 15545-50. doi:10.1073/pnas.0506580102. PMC PMC1239896. PMID 16199517. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1239896.
- ↑ Draghici, S.; Khatri, P.; Laurentiu, A. et al. (2007). "A systems biology approach for pathway level analysis". Genome Research 17 (10): 1537–45. doi:10.1101/gr.6202607. PMC PMC1987343. PMID 17785539. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1987343.
- ↑ Ősz, Á.; Pongor, L.S.; Szirmai, D. et al. (2019). "A snapshot of 3649 Web-based services published between 1994 and 2017 shows a decrease in availability after two years". Briefings in Bioingformatics 20 (3): 1004-1010. doi:10.1093/bib/bbx159. PMC PMC6585384. PMID 29228189. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6585384.
- ↑ Yu, G.; Wang,L.-G.; Hanf, Y. et al. (2012). "clusterProfiler: an R package for comparing biological themes among gene clusters". OMICS 16 (5): 284–7. doi:10.1089/omi.2011.0118. PMC PMC3339379. PMID 22455463. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3339379.
- ↑ Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D. et al. (2016). "Enrichr: A comprehensive gene set enrichment analysis web server 2016 update". Nucleic Acids Research 44 (W1): W90—7. doi:10.1093/nar/gkw377. PMC PMC4987924. PMID 27141961. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4987924.
- ↑ Young, M.D.; Wakefield, M.J.; Smyth, G.K. et al. (2010). "Gene ontology analysis for RNA-seq: accounting for selection bias". Genome Biology 11 (2): R14. doi:10.1186/gb-2010-11-2-r14. PMC PMC2872874. PMID 20132535. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2872874.
- ↑ Bindea, G.; Mlecnik, B.; Hackl, H. et al. (2009). "ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks". Bioinformatics 25 (8): 1091–3. doi:10.1093/bioinformatics/btp101. PMC PMC2666812. PMID 19237447. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2666812.
- ↑ Giannoulatouet, E.; Park, S.-H.; Humphreys, D.T. al. (2014). "Verification and validation of bioinformatics software without a gold standard: A case study of BWA and Bowtie". BMC Bioinformatics 15 (Suppl. 16): S15. doi:10.1186/1471-2105-15-S16-S15. PMC PMC4290646. PMID 25521810. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4290646.
- ↑ Curtis, R.K.; Oresic, M.; Vidal-Puig, A. (2005). "Pathways to the analysis of microarray data". Trends in Biotechnology 23 (8): 429-35. doi:10.1016/j.tibtech.2005.05.011. PMID 15950303.
- ↑ Tarca, A.L.; Draghici, S.; Bhatti, G. et al. (2012). "Down-weighting overlapping genes improves gene set analysis". BMC Bioinformatics 13: 136. doi:10.1186/1471-2105-13-136. PMC PMC3443069. PMID 22713124. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3443069.
- ↑ Tomfohr, J.; Lu, J.; Kepler, T.B. (2005). "Pathway level analysis of gene expression using singular value decomposition". BMC Bioinformatics 6: 225. doi:10.1186/1471-2105-6-225. PMC PMC1261155. PMID 16156896. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1261155.
- ↑ Tarca, A.L.; Draghici, S.; Khatri, P. et al. (2009). "A novel signaling pathway impact analysis". Bioinformatics 25 (1): 75–82. doi:10.1093/bioinformatics/btn577. PMC PMC2732297. PMID 18990722. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2732297.
- ↑ Naeem, H.; Zimmer, R.; Tavakkolkhah, P. et al. (2012). "Rigorous assessment of gene set enrichment tests". Bioinformatics 28 (11): 1480-6. doi:10.1093/bioinformatics/bts164. PMID 22492315.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation, spelling, and grammar. In some cases important information was missing from the references, and that information was added.