Difference between revisions of "Journal:Principles of metadata organization at the ENCODE data coordination center"
Shawndouglas (talk | contribs) (Saving and adding more content.) |
Shawndouglas (talk | contribs) (Saving and adding more content.) |
||
| Line 136: | Line 136: | ||
|} | |} | ||
|} | |} | ||
The ENCODE metadata data model is a hybrid relational-object-based data store in which the major categories of metadata are represented as one or more JSON objects implemented in JSON-SCHEMA and JSON-LD.<ref name="HitzSnoVault16">{{cite journal |title=SnoVault and encodeD: A novel object-based storage system and applications to ENCODE metadata |journal=bioRxiv |author=Hitz, B.; Rowe, L.D.; Podduturi, N. et al. |year=2016 |doi=10.1101/044578 |quote=This article is a preprint and has not been peer-reviewed.}}</ref> Because different aspects of experimental and computational assays are reused, each category of metadata that represents an experimental variable can be referred to multiple times (Figure 4). For example, a single donor can contribute multiple biosamples, such as a liver and brain, or multiple assays can use the same tissue. | |||
[[File:Fig4 Hong Database2016 2016.jpg|756px]] | |||
{{clear}} | |||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| border="0" cellpadding="5" cellspacing="0" width="756px" | |||
|- | |||
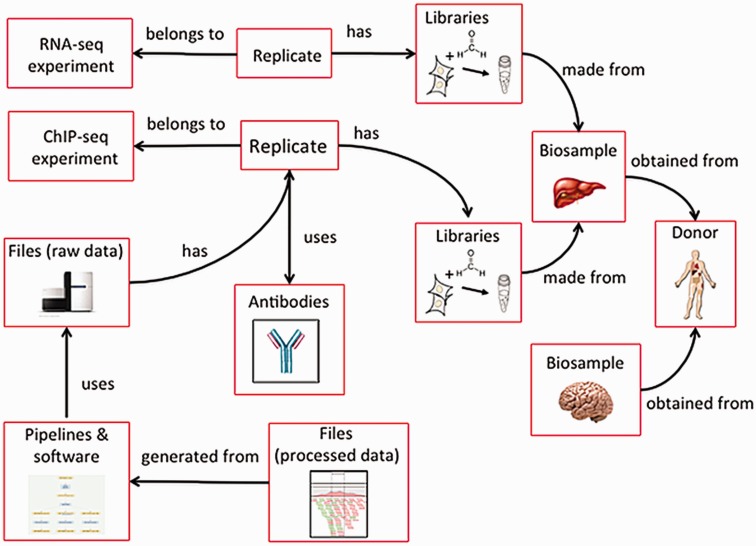
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 4.''' Categories in the metadata model are linked to each other. Categories of metadata are linked to each other and can be described by relationships between the categories. Each individual category can be referred to multiple times. For example, a liver and a brain can be obtained from the same donor. In addition, a single biosample, like the liver, can be used as input for multiple assays. Because each donor and biosample is accessioned, they can be referred to uniquely.</blockquote> | |||
|- | |||
|} | |||
|} | |||
The detailed metadata are captured as distinct structured fields or in protocol documents associated with the experiment. The decision as to whether the metadata are highlighted as a separate field is reflective of whether that specific metadata fulfills a set of the five criteria described above. | |||
===Curation and validation of metadata=== | |||
Use of controlled vocabularies and ontologies ensures consistent description of the metadata. The ENCODE DCC uses the appropriate ontologies, when available, that contain defined relationships used to capture the values for the metadata.<ref name="MalladiOntology15">{{cite journal |title=Ontology application and use at the ENCODE DCC |journal=Database |author=Malladi, V.S.; Erickson, D.T.; Podduturi, N.R. et al. |volume=2015 |pages=bav010 |year=2015 |doi=10.1093/database/bav010 |pmid=25776021 |pmc=PMC4360730}}</ref> These include the description of what the biosample is: UBERON for tissues, CL for primary cell types and EFO for immortalized cell lines.<ref name="MungallUberon12">{{cite journal |title=Uberon, an integrative multi-species anatomy ontology |journal=Genome Biology |author=Mungall, C.J.; Torniai, C.; Gkoutos, G.V. et al. |volume=13 |issue=1 |pages=R5 |year=2012 |doi=10.1186/gb-2012-13-1-r5 |pmid=22293552 |pmc=PMC3334586}}</ref><ref name="BardAnOnt05">{{cite journal |title=An ontology for cell types |journal=Genome Biology |author=Bard, J.; Rhee, S.Y.; Ashburner, M. |volume=6 |issue=2 |pages=R21 |year=2005 |doi=10.1186/gb-2005-6-2-r21 |pmid=15693950 |pmc=PMC551541}}</ref><ref name="MaloneModeling10">{{cite journal |title=Modeling sample variables with an Experimental Factor Ontology |journal=Bioinformatics |author=Malone, J.; Holloway, E.; Adamusiak, T. et al. |volume=26 |issue=8 |pages=1112-8 |year=2010 |doi=10.1093/bioinformatics/btq099 |pmid=20200009 |pmc=PMC2853691}}</ref> For treatments on the biosample, ChEBI terms will be used.<ref name="HastingsTheChEBI13">{{cite journal |title=The ChEBI reference database and ontology for biologically relevant chemistry: Enhancements for 2013 |journal=Nucleic Acids Research |author=Hastings, J.; de Matos, P.; Dekker, A. et al. |volume=41 |issue=D1 |pages=D456-63 |year=2013 |doi=10.1093/nar/gks1146 |pmid=23180789 |pmc=PMC3531142}}</ref> OBI is used for capturing the assay name.<ref name="BrinkmanModeling10">{{cite journal |title=Modeling biomedical experimental processes with OBI |journal=Journal of Biomedical Semantics |author=Brinkman, R.R.; Courtot, M.; Derom, D. et al. |volume=1 |issue=Suppl 1 |pages=S7 |year=2010 |doi=10.1186/2041-1480-1-S1-S7 |pmid=20626927 |pmc=PMC2903726}}</ref> The use of ontologies allows instant interoperability with other datasets that use the same ontologies.<ref name="MalladiOntology15" /> When no ontology is available, a controlled list of terms is provided in the schema as an enumerated list order to maintain rigorous consistency (Figure 5). The use of ontologies is more significant when multiple projects agree on their use as this allows interoperability between projects empowering greater use of their results. | |||
| Line 142: | Line 161: | ||
==Notes== | ==Notes== | ||
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. | This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. An additional reference was added, originally referenced as "Hitz ''et al.'', in preparation; the pre-print non-peer-reviewed version is now referenced for the sake of context. | ||
<!--Place all category tags here--> | <!--Place all category tags here--> | ||
Revision as of 21:58, 6 September 2016
| Full article title | Principles of metadata organization at the ENCODE data coordination center |
|---|---|
| Journal | Database |
| Author(s) |
Hong, Eurie L.; Sloan, Cricket A.; Chan, Esther T.; Davidson, Jean M.; Malladi, Venkat S.; Strattan, J. Seth; Hitz, Benjamin C.; Gabdank, Idan; Narayanan, Aditi K.; Ho, Marcus; Lee, Brian T.; Rowe, Laurence D.; Dreszer, Timothy R.; Roe, Greg R.; Podduturi, Nikhil R.; Tanaka, Forrest; Hilton, Jason A.; Cherry, J. Michael |
| Author affiliation(s) | Stanford University, University of California - Santa Cruz |
| Primary contact | Email: cherry at stanford dot edu |
| Year published | 2016 |
| Page(s) | baw001 |
| DOI | 10.1093/database/baw001 |
| ISSN | 1758-0463 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | http://database.oxfordjournals.org/content/2016/baw001 |
| Download | http://database.oxfordjournals.org/content/2016/baw001.full.pdf+html (PDF) |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
The Encyclopedia of DNA Elements (ENCODE) Data Coordinating Center (DCC) is responsible for organizing, describing and providing access to the diverse data generated by the ENCODE project. The description of these data, known as metadata, includes the biological sample used as input, the protocols and assays performed on these samples, the data files generated from the results and the computational methods used to analyze the data. Here, we outline the principles and philosophy used to define the ENCODE metadata in order to create a metadata standard that can be applied to diverse assays and multiple genomic projects. In addition, we present how the data are validated and used by the ENCODE DCC in creating the ENCODE Portal (https://www.encodeproject.org/).
Database URL: www.encodeproject.org
Introduction
The goal of the Encyclopedia of DNA Elements (ENCODE) project is to annotate functional regions in the human and mouse genomes. Functional regions include those that code protein-coding or non-coding RNA gene products as well as regions that could have a regulatory role.[1][2] To this end, the project has surveyed the landscape of the human genome using over 35 high-throughput experimental methods in more than 250 different cell and tissue types, resulting in over 4000 experiments.[1][3] These datasets are submitted to a Data Coordinating Center (DCC), whose role is to describe, organize and provide access to these diverse datasets.[4]
A description of these datasets, collectively known as metadata, encompasses, but is not limited to, the identification of the experimental method used to generate the data, the sex and age of the donor from whom a skin biopsy was taken, and the software used to align the sequencing reads to a reference genome. Defining and organizing the set of metadata that is relevant, informative and applicable to diverse experimental techniques is challenging. These challenges are not unique to the ENCODE DCC. Several major experimental consortia similar in scale to the ENCODE project exist, as well as public database projects that collect and distribute high-throughput genomic data. Analogous to the ENCODE project, the modENCODE project was begun in 2007 to identify functional elements in the model organisms Caenorhabditis elegans and Drosophila melanogaster. The modENCODE DCC faced similar challenges in trying to integrate diverse data types using a variety of experimental techniques.[5] Other consortia, such as the Roadmap Epigenomics Mapping Centers, also have been tasked with defining the metadata.[6] In addition, databases such as ArrayExpress at the EBI, GEO and SRA at the NCBI, Data Dryad (http://datadryad.org/) and FigShare (http://figshare.com/) serve as data repositories, accepting diverse data types from large consortia as well as from individual research laboratories.[7][8][9]
The challenges of capturing metadata and organizing high-throughput genomic datasets are not unique to NIH-funded consortia and data repositories. Since many researchers submit their high-throughput data to data repositories and scientific data publications, tools and data management software, such as the Investigation Assay Study tools (ISA-tools) and laboratory information management systems (LIMS), provide resources aimed to help laboratories organize their data for better compatibility with these data repositories.[10] In addition, there have been multiple efforts to define a minimal set of metadata for genomic assays, including standards proposed by the Functional Genomics Data society (FGED; http://fged.org/projects/minseqe/) and the Global Alliance for Genomics and Health (GA4GH; https://github.com/ga4gh/schemas), to improve interoperability among data generated by diverse groups.
Here, we describe how metadata are organized at the ENCODE DCC and define the metadata standard that is used to describe the experimental assays and computational analyses generated by the ENCODE project. The metadata standard includes the principles driving the selection of metadata as well as how these metadata are validated and used by the DCC. Understanding the principles and data organization will help improve the accessibility of the ENCODE datasets as well as provide transparency to the data generation processes. This understanding will allow integration of the diverse data within the ENCODE consortium as well as integration with related assays from other large-scale consortium projects and individual labs.
Metadata describing ENCODE assays
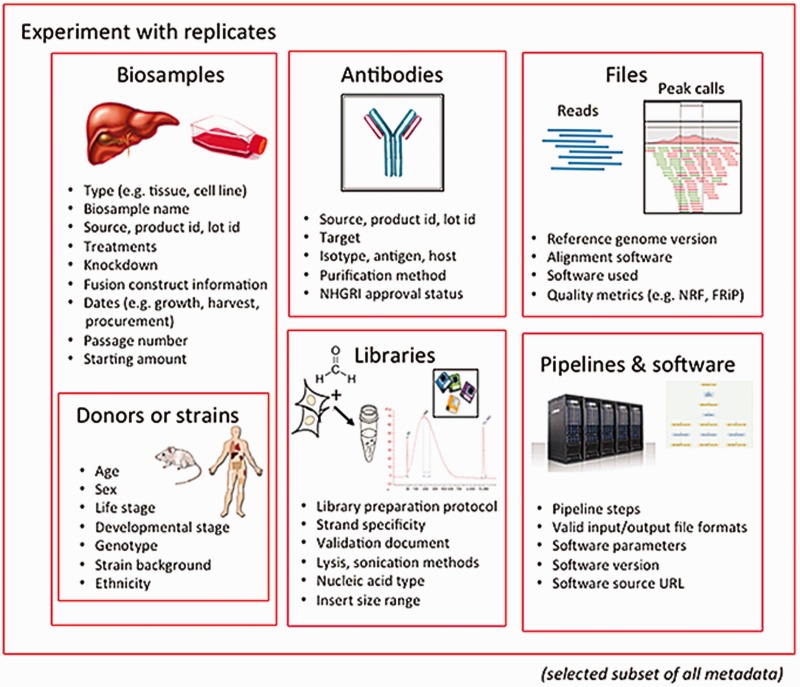
The categories of metadata currently being collected by the ENCODE DCC builds on the set collected during the previous phases of the project. During the earlier phases, a core set of metadata describing the assays, cell types and antibodies were submitted to the ENCODE DCC.[11] The current metadata set expands the number of categories into the following major organizational units: biosamples, libraries, antibodies, experiments, data files and pipelines (Figure 1). Only a selected set of metadata are included below as examples, to give a sense of the breadth and depth of our approach.
|
The biological material used as input material for an experimental assay is called a biosample. This category of metadata is an expansion of the ‘cell types’ captured in previous phases of the ENCODE project.[11] Biosample metadata includes non-identifying information about the donors (if the sample is from a human) and details of strain backgrounds (if the sample is from model organisms) (Figure 1). Metadata for the biosample includes the source of the material (such as a company name or a lab), how it was handled in the lab (such as number of passages or starting amounts) and any modifications to the biological material (such as the integration of a fusion gene or the application of a treatment).
The library refers to the nucleic acid material that is extracted from the biosample and contains details of the experimental methods used to prepare that nucleic acid for sequencing. Details of the specific population or sub-population of nucleic acid (e.g. DNA, rRNA, nuclear RNA, etc.) and how this material is prepared for sequencing libraries is captured as metadata.
The metadata recorded for antibodies include the source of the antibody, as well as the product number and the specific lot of the antibody if acquired commercially. Capturing the antibody lot id is critical because there is potential for lot-to-lot variation in the specificity and sensitivity of an antibody. Antibody metadata include characterizations of the antibody performed by the labs, which examines this specificity and sensitivity of an antibody, as defined by the ENCODE consortium.[12]
The experiment refers to one or more replicates that are grouped together along with the raw data files and processed data files. Each replicate that is part of an experiment will be performed using the same experimental method or assay (e.g. ChIP-seq). A single replicate, which can be designated as a biological or technical replicate, is linked to a specific library and an antibody used in immunoprecipitation-based assay (e.g. ChIP-seq). Since the library is derived from the biosample, the details of the biosample are affiliated with the replicate through the library used.
A single experiment can include multiple files. These files include, but are not limited to, the raw data (typically sequence reads), the mapping of these sequence reads against the reference genome, and genomic features that are represented by these reads (often called ‘signals’ or ‘peaks’). Metadata pertaining to files include the file format and a short description, known as an output type, of the contents of the file.
In addition to capturing the format of the files generated (e.g. fastq, BAM, bigWig), metadata regarding quality control metrics, the software, the version of the software and pipelines used to generate the file are included as part of the pipeline-related set of metadata. The metadata for a given file also consist of other files that are connected through input and output relationships.
Defining the metadata standard
The categories of metadata captured for the ENCODE experiments described in the previous section aim to provide a summary of the experimental and computational methods as well as provide enough information to facilitate the evaluation and interpretation of multiple experiments by the scientific community. The breadth and depth of metadata selected in each of the categories need to be able to uniquely identify experiments from each other. The following five principles guide the selection of the specific metadata in each category.
Principle 1: Reflect experimental variables
The detailed metadata in each category are selected to reflect potential experimental variables when evaluating a set of similar assays. The key metadata may differ depending on the assays examined by the researcher so the set of metadata included for the ENCODE assays strives to be broadly applicable to multiple assays without sacrificing specificity for a single assay. To this end, extensive metadata describing the biosamples used in assays, preparation of the libraries and software used to generate the files are captured. Selected examples highlighting this principle are described.
The biosample metadata category includes donor information for human samples, such as the age and sex, or strain background information for model organisms, such as strain names and genotypes. Whether the biosample has been treated with a chemical or biological agent (such as tamoxifen or an infectious agent), contains a fusion protein (such as an eGFP tagged protein), or has been transfected with an RNAi to knock-down protein levels, also will be reflected in the biosample metadata category.
Preparation of libraries can dictate and influence which sets of data can be appropriately compared together, as well as influence the types of software that can be used to analyze the data. In particular, the processing of RNA-seq data differs on the length of the RNA population being selected, whether the RNA contains a 5′ 7-methylguanosine cap, and whether rRNA populations were removed prior to library prep. Although many of these details are not generally applicable to other assays, they are included in the ENCODE metadata set because they are essential in understanding the experimental variables for an RNA-seq assay.
The contents of a data file are strongly dependent on the software used to generate the file. Software with similar functions, such as an aligner, can produce different results. In addition, different parameters passed into a same software can result in different output. Therefore, file metadata category includes software information, consisting of version numbers, md5sum of the downloaded software and supporting documentation regarding parameters used.
Principle 2: Help uniquely identify re-usable reagents
In addition to reflecting experimental variables, the metadata can provide sufficient information to differentiate between similar entities that are often used as reagents. The ability to distinguish between similar but not identical reagents is essential to ensure a specific biosample is used as input for different assays or the same lot of antibody is used for multiple ChIP-seq assays.
For example, tissues and primary cells can be differentiated by a difference between donors. Furthermore, identical biosamples from an individual donor, such as a blood draw, can be distinguished by recoding dates of collection for tissues and primary cell lines. Cell lines, whether immortalized or differentiated in vitro, can be distinguished as unique growths based on the date the batch was started. For antibodies, the lot number of the antibody is captured in addition to the vendor and product ID due to potential variation between lots.
Principle 3: Encourage reproducibility and interpretation of the data
Another core principle of defining the ENCODE metadata standard is to include essential experimental and computational details that allow researchers to repeat the assays or analysis as well as provide insight and context to evaluate the quality of the experiment. For biosamples, the amount of the starting material used (in number of cells or weight) is included. In addition, metadata such as the source, including any product numbers and lot IDs, will be recorded to allow other members of the scientific community to obtain similar biosamples. For libraries, this includes the methods used to lyse and prepare the nucleic acid for sequencing. For files, the versions of the software and the pipeline used are captured as well as the input files and reference files used to generate the output files of the analysis. Capturing this level of experimental detail allows easier comparison between different assays, computational results and analyses.
Principle 4: Represent data standards
The ENCODE Consortium has defined standards for a range of different aspects of the experimental and analysis process (https://www.encodeproject.org/data-standards/). The data standards include how assays should be performed as well as how data results should be analyzed. For example, there are standards agreed upon by the consortium for the consistent treatment of cells, including growth protocols and the number of passages. Standards describe how to evaluate the specificity and sensitivity of that antibody against the target have also been developed.[12] In addition, standards for read depth and analysis methods are also agreed upon by the ENCODE Consortium. Since these are significant details about the assay as defined by the consortium, these specific metadata are included in the ENCODE metadata set. Growth protocols and other protocols for preparing the biosample and libraries are included as documents. Quality control metrics on read depth, uniquely mapping reads and replicate concordance values are captured as well.
Principle 5: Facilitate searching and identification of experimental datasets
The metadata are used for searching data generated by the ENCODE Consortium at the ENCODE Portal (https://www.encodeproject.org/). In addition to previously mentioned metadata, other metadata were selected that can help improve searching and identification of the datasets of interest. For example, any set of files or assay can be associated with a citation in order to easily find data used in a given publication. In addition, the lab submitting the generated data is included to allow searching for data generated by a specific lab.
Implementing the metadata principles
Accessions
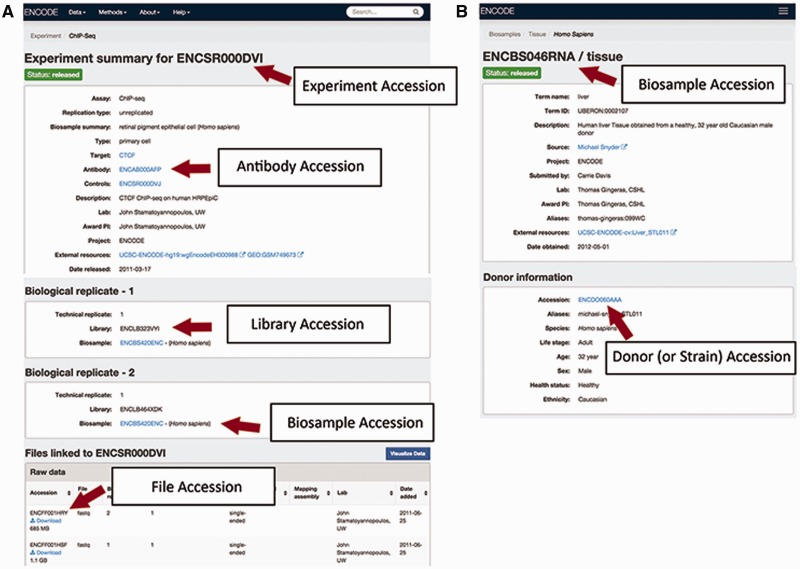
Within each of the categories of metadata, the uniquely identified experimental variables can help distinguish similar entities. In order to easily refer to these entities in data submission as well as in publications, ENCODE accessions are assigned to key metadata categories. Each ENCODE accession is stable and will be tracked once they are created.
The accessions are in the format ENC[SR|BS |DO|AB| LB|FF] [0-9]{3} [A-Z]{3} where [SR|BS| DO|AB|LB |FF] refer to the metadata type given the accession (Figure 2). This allows for more than 17 million accessions to be generated by entity type.
|
Accessions are given to the following types of metadata:
An experiment: An experiment accession refers to one or more replicates that are grouped together along with the raw data files and processed data files. Typically, each replicate will be performed using the same method, performed on the same kind of biosample and investigating the same target (see the "Creating the ENCODE metadata data model" section for more details). A sample accession for an assay is ENCSR000DVI (Figure 2A).
A biosample: An accessioned biosample refers to a tube or sample of that biological material that is being used. For example, the following would all be given a biosample accession: (1) a batch of a cell line grown on a specific day, (2) the isolation of a primary cell culture on a specific day or (3) the dissection of a tissue sample on a specific day. An example of biosample accession is ENCBS046RNA (Figure 2B).
A strain or donor: Every strain background (for model organisms) and donor (for humans) is given a donor accession. This accession allows multiple samples obtained from a single donor to be grouped together. The donor information is listed within the biosample, for example in ENCBS046RNA (Figure 2B).
An antibody lot: Each unique combination of an antibody’s source, production number and lot is accessioned so that assays can refer specifically to that antibody. Each antibody lot is also associated with characterizations for its target in a species, for example in ENCAB934MDN (Figure 2A).
A library: A unique library that was sequenced is accessioned to ensure that the correct files are associated with the nucleic acid material that has been created from the biosample. The library accession and experimental details of how the library was constructed are displayed on the assay page, i.e. ENCSR000DVI (Figure 2A).
A file: Each data, analysis and reference file are accessioned. This accession is used as the file name, along with its file format as an extension. The file accession is associated with the contents of that file. When a new file is submitted to replace an existing file, the new file is given a new accession and related to the older file. Files are displayed at the bottom of an assay page, i.e. ENCSR000DVI (Figure 2A).
Creating the ENCODE metadata data model
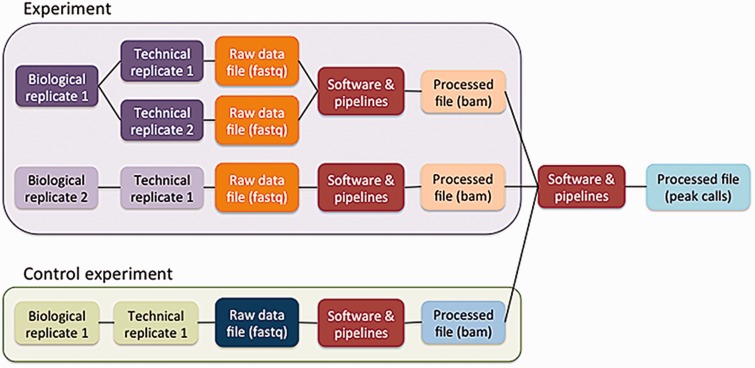
The five principles are embodied in the ENCODE metadata model (https://github.com/ENCODE-DCC/encoded/tree/master/src/encoded/schemas or https://www.encodeproject.org/profiles/). The metadata are the details collected about the experiments and reagents, the metadata model is the definition of what can be collected, and the metadata data model is the computational structure used to store and organize the metadata model. The major categories of metadata are organized in a model that reflects how researchers perform the experimental and computational assays (Figure 3). Experiments in the metadata model can contain one or more replicates, representing the number of times an assay was performed in an attempt to demonstrate reproducibility. These repetitions can be classified as biological or technical replicates. Biological replicates, representing libraries made from distinct biosamples, can be further specified to indicate whether that biosample was derived from the same donor (isogenic) or from different donors (anisogenic). Technical replicates are linked to the same biosample. Although the ENCODE Consortium defines technical replicates as two different libraries of nucleic acid prepared from the same biosample, the metadata model can accommodate alternate definitions. Each replicate (either biological or technical) is linked to raw data files that are used in pipelines to generate additional processed files.
|
The ENCODE metadata data model is a hybrid relational-object-based data store in which the major categories of metadata are represented as one or more JSON objects implemented in JSON-SCHEMA and JSON-LD.[13] Because different aspects of experimental and computational assays are reused, each category of metadata that represents an experimental variable can be referred to multiple times (Figure 4). For example, a single donor can contribute multiple biosamples, such as a liver and brain, or multiple assays can use the same tissue.
|
The detailed metadata are captured as distinct structured fields or in protocol documents associated with the experiment. The decision as to whether the metadata are highlighted as a separate field is reflective of whether that specific metadata fulfills a set of the five criteria described above.
Curation and validation of metadata
Use of controlled vocabularies and ontologies ensures consistent description of the metadata. The ENCODE DCC uses the appropriate ontologies, when available, that contain defined relationships used to capture the values for the metadata.[14] These include the description of what the biosample is: UBERON for tissues, CL for primary cell types and EFO for immortalized cell lines.[15][16][17] For treatments on the biosample, ChEBI terms will be used.[18] OBI is used for capturing the assay name.[19] The use of ontologies allows instant interoperability with other datasets that use the same ontologies.[14] When no ontology is available, a controlled list of terms is provided in the schema as an enumerated list order to maintain rigorous consistency (Figure 5). The use of ontologies is more significant when multiple projects agree on their use as this allows interoperability between projects empowering greater use of their results.
References
- ↑ 1.0 1.1 ENCODE Project Consortium (2012). "An integrated encyclopedia of DNA elements in the human genome". Nature 489 (7414): 57-74. doi:10.1038/nature11247. PMC PMC3439153. PMID 22955616. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3439153.
- ↑ Yue, F.; Cheng, Y.; Breschi, A. et al. (2014). "A comparative encyclopedia of DNA elements in the mouse genome". Nature 515 (7527): 355-64. doi:10.1038/nature13992. PMC PMC4266106. PMID 25409824. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4266106.
- ↑ ENCODE Project Consortium et al. (2007). "Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project". Nature 447 (7146): 799–816. doi:10.1038/nature05874. PMC PMC2212820. PMID 17571346. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2212820.
- ↑ Sloan, C.A.; Chan, E.T.; Davidson, J.M. et al. (2016). "ENCODE data at the ENCODE portal". Nucleic Acids Research 44 (D1): D726-32. doi:10.1093/nar/gkv1160. PMC PMC4702836. PMID 26527727. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4702836.
- ↑ Washington, N.L.; Stinson, E.O.; Perry, M.D. et al. (2011). "The modENCODE Data Coordination Center: Lessons in harvesting comprehensive experimental details". Database 2011: bar023. doi:10.1093/database/bar023. PMC PMC3170170. PMID 21856757. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3170170.
- ↑ Bernstein, B.E.; Stamatoyannopoulos, J.A.; Costello, J.F. et al. (2010). "The NIH Roadmap Epigenomics Mapping Consortium". Nature Biotechnology 28 (10): 1045-8. doi:10.1038/nbt1010-1045. PMC PMC3607281. PMID 20944595. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3607281.
- ↑ Kolesnikov, N.; Hastings, E.; Keays, M. et al. (2015). "ArrayExpress update -- Simplifying data submissions". Nucleic Acids Research 43 (D1): D1113-6. doi:10.1093/nar/gku1057. PMC PMC4383899. PMID 25361974. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4383899.
- ↑ Barrett, T.; Wilhite, S.E.; Ledoux, P. et al. (2013). "NCBI GEO: Archive for functional genomics data sets -- Update". Nucleic Acids Research 41 (D1): D991-5. doi:10.1093/nar/gks1193. PMC PMC3531084. PMID 23193258. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3531084.
- ↑ NCBI Resource Coordinators (2015). "Database resources of the National Center for Biotechnology Information". Nucleic Acids Research 43 (D1): D6–17. doi:10.1093/nar/gku1130. PMC PMC4383943. PMID 25398906. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4383943.
- ↑ Rocca-Serra, P.; Brandizi, M.; Maquire, E. et al. (2010). "ISA software suite: Supporting standards-compliant experimental annotation and enabling curation at the community level". Bioinformatics 26 (18): 2354-6. doi:10.1093/bioinformatics/btq415. PMC PMC2935443. PMID 20679334. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2935443.
- ↑ 11.0 11.1 Rosenbloom, K.R.; Dreszer, T.R.; Long, J.C. et al. (2012). "ENCODE whole-genome data in the UCSC Genome Browser: Update 2012". Nucleic Acids Research 40 (D1): D912-7. doi:10.1093/nar/gkr1012. PMC PMC3245183. PMID 22075998. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3245183.
- ↑ 12.0 12.1 Landt, S.G.; Marinov, G.K.; Kundaje, A. et al. (2012). "ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia". Genome Research 22 (9): 1813-31. doi:10.1101/gr.136184.111. PMC PMC3431496. PMID 22955991. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3431496.
- ↑ Hitz, B.; Rowe, L.D.; Podduturi, N. et al. (2016). "SnoVault and encodeD: A novel object-based storage system and applications to ENCODE metadata". bioRxiv. doi:10.1101/044578. "This article is a preprint and has not been peer-reviewed."
- ↑ 14.0 14.1 Malladi, V.S.; Erickson, D.T.; Podduturi, N.R. et al. (2015). "Ontology application and use at the ENCODE DCC". Database 2015: bav010. doi:10.1093/database/bav010. PMC PMC4360730. PMID 25776021. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4360730.
- ↑ Mungall, C.J.; Torniai, C.; Gkoutos, G.V. et al. (2012). "Uberon, an integrative multi-species anatomy ontology". Genome Biology 13 (1): R5. doi:10.1186/gb-2012-13-1-r5. PMC PMC3334586. PMID 22293552. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3334586.
- ↑ Bard, J.; Rhee, S.Y.; Ashburner, M. (2005). "An ontology for cell types". Genome Biology 6 (2): R21. doi:10.1186/gb-2005-6-2-r21. PMC PMC551541. PMID 15693950. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC551541.
- ↑ Malone, J.; Holloway, E.; Adamusiak, T. et al. (2010). "Modeling sample variables with an Experimental Factor Ontology". Bioinformatics 26 (8): 1112-8. doi:10.1093/bioinformatics/btq099. PMC PMC2853691. PMID 20200009. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2853691.
- ↑ Hastings, J.; de Matos, P.; Dekker, A. et al. (2013). "The ChEBI reference database and ontology for biologically relevant chemistry: Enhancements for 2013". Nucleic Acids Research 41 (D1): D456-63. doi:10.1093/nar/gks1146. PMC PMC3531142. PMID 23180789. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3531142.
- ↑ Brinkman, R.R.; Courtot, M.; Derom, D. et al. (2010). "Modeling biomedical experimental processes with OBI". Journal of Biomedical Semantics 1 (Suppl 1): S7. doi:10.1186/2041-1480-1-S1-S7. PMC PMC2903726. PMID 20626927. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2903726.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. An additional reference was added, originally referenced as "Hitz et al., in preparation; the pre-print non-peer-reviewed version is now referenced for the sake of context.