Difference between revisions of "Journal:Semantics for an integrative and immersive pipeline combining visualization and analysis of molecular data"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) (Saving and adding more.) |
||
| Line 104: | Line 104: | ||
On the application side, the use of ontologies in order to standardize knowledge in scientific fields underwent an important and spontaneous growth at the end of the 1990s.<ref name="Schulze-KremerOnto02">{{cite journal |title=Ontologies for molecular biology and bioinformatics |journal=In Silico Biology |author=Schulze-Kremer, S. |volume=2 |issue=3 |pages=179–93 |year=2002 |pmid=12542404}}</ref> Bioinformatics, tightly anchored in structural biology, has used ontologies for a long time. The most significant example is the fast-growing [[Genomics|genomic field]], in which it became impossible to handle data flow without a proper and standardized organization of the data.<ref name="SchuurmanOnto08">{{cite journal |title=Ontologies for bioinformatics |journal=Bioinformatics and Biology Insights |author=Schuurman, N.; Leszcynski, A. |volume=2 |pages=187—200 |year=2008 |pmid=19812775 |pmc=PMC2735951}}</ref> The tool Gene Ontology<ref name="GOCGene00">{{cite journal |title=Gene ontology: Tool for the unification of biology |journal=Nature Genetics |author=The Gene Ontology Consortium, Ashburner, M.; Ball, C.A. et al. |volume=25 |issue=1 |pages=25–9 |year=2000 |doi=10.1038/75556 |pmid=10802651 |pmc=PMC3037419}}</ref> regroups genomic data into a uniform format and a knowledge base. Currently, it is one of the most referred to ontologies in the literature. Rabattu ''et al.''<ref name="RabattuMyCorporis15">{{cite journal |title=My Corporis Fabrica Embryo: An ontology-based 3D spatio-temporal modeling of human embryo development |journal=Journal of Biomedical Semantics |author=Rabattu, P.Y.; Massé, B.; Ulliana, F. et al. |volume=6 |pages=36 |year=2015 |doi=10.1186/s13326-015-0034-0 |pmid=26413258 |pmc=PMC4582726}}</ref> propose an approach to spatio-temporal reasoning on semantic descriptions of an evolving human embryo. Several biological databases or organizations such as UniProtKB1 and the Open Biomedical Ontologies<ref name="SmithTheOBO07">{{cite journal |title=The OBO Foundry: Coordinated evolution of ontologies to support biomedical data integration |journal=Nature Biotechnology |author=Smith, B.; Ashburner, M.; Rosse, C. et al. |volume=25 |issue=11 |pages=1251–5 |year=2007 |doi=10.1038/nbt1346 |pmid=17989687 |pmc=PMC2814061}}</ref> provide ways to access data or ontologies under RDF or OWL format to allow their use in expert tools or specific pipelines. One can also note the open-source project Bio2RDF<ref name="BelleauBio2RDF">{{cite journal |title=Bio2RDF: towards a mashup to build bioinformatics knowledge systems |journal=Journal of Biomedical Informatics |author=Belleau, F.; Nolin, M.A;. Tourigny, N. et al. |volume=41 |issue=5 |pages=706–16 |year=2008 |doi=10.1016/j.jbi.2008.03.004 |pmid=18472304}}</ref> that aims to build and provide the largest network of "Linked Data for the Life Sciences" using semantic web approaches. | On the application side, the use of ontologies in order to standardize knowledge in scientific fields underwent an important and spontaneous growth at the end of the 1990s.<ref name="Schulze-KremerOnto02">{{cite journal |title=Ontologies for molecular biology and bioinformatics |journal=In Silico Biology |author=Schulze-Kremer, S. |volume=2 |issue=3 |pages=179–93 |year=2002 |pmid=12542404}}</ref> Bioinformatics, tightly anchored in structural biology, has used ontologies for a long time. The most significant example is the fast-growing [[Genomics|genomic field]], in which it became impossible to handle data flow without a proper and standardized organization of the data.<ref name="SchuurmanOnto08">{{cite journal |title=Ontologies for bioinformatics |journal=Bioinformatics and Biology Insights |author=Schuurman, N.; Leszcynski, A. |volume=2 |pages=187—200 |year=2008 |pmid=19812775 |pmc=PMC2735951}}</ref> The tool Gene Ontology<ref name="GOCGene00">{{cite journal |title=Gene ontology: Tool for the unification of biology |journal=Nature Genetics |author=The Gene Ontology Consortium, Ashburner, M.; Ball, C.A. et al. |volume=25 |issue=1 |pages=25–9 |year=2000 |doi=10.1038/75556 |pmid=10802651 |pmc=PMC3037419}}</ref> regroups genomic data into a uniform format and a knowledge base. Currently, it is one of the most referred to ontologies in the literature. Rabattu ''et al.''<ref name="RabattuMyCorporis15">{{cite journal |title=My Corporis Fabrica Embryo: An ontology-based 3D spatio-temporal modeling of human embryo development |journal=Journal of Biomedical Semantics |author=Rabattu, P.Y.; Massé, B.; Ulliana, F. et al. |volume=6 |pages=36 |year=2015 |doi=10.1186/s13326-015-0034-0 |pmid=26413258 |pmc=PMC4582726}}</ref> propose an approach to spatio-temporal reasoning on semantic descriptions of an evolving human embryo. Several biological databases or organizations such as UniProtKB1 and the Open Biomedical Ontologies<ref name="SmithTheOBO07">{{cite journal |title=The OBO Foundry: Coordinated evolution of ontologies to support biomedical data integration |journal=Nature Biotechnology |author=Smith, B.; Ashburner, M.; Rosse, C. et al. |volume=25 |issue=11 |pages=1251–5 |year=2007 |doi=10.1038/nbt1346 |pmid=17989687 |pmc=PMC2814061}}</ref> provide ways to access data or ontologies under RDF or OWL format to allow their use in expert tools or specific pipelines. One can also note the open-source project Bio2RDF<ref name="BelleauBio2RDF">{{cite journal |title=Bio2RDF: towards a mashup to build bioinformatics knowledge systems |journal=Journal of Biomedical Informatics |author=Belleau, F.; Nolin, M.A;. Tourigny, N. et al. |volume=41 |issue=5 |pages=706–16 |year=2008 |doi=10.1016/j.jbi.2008.03.004 |pmid=18472304}}</ref> that aims to build and provide the largest network of "Linked Data for the Life Sciences" using semantic web approaches. | ||
Only a few expert software packages based on ontologies have been developed for structural biology. Avogadro<ref name="HanwellAvo12">{{cite journal |title=Avogadro: An advanced semantic chemical editor, visualization, and analysis platform |journal=Journal of Cheminformatics |author=Hanwell, M.D.; Curtis, D.E.; Lonie, D.C. et al. |volume=4 |issue=1 |pages=17 |year=2012 |doi=10.1186/1758-2946-4-17 |pmid=22889332 |pmc=PMC3542060}}</ref> and DIVE<ref name="RysavyDIVE14">{{cite journal |title=DIVE: A Graph-Based Visual-Analytics Framework for Big Data |journal=IEEE Computer Graphics and Applications |author=Rysavy, S.J.; Bromley, D.; Daggett, V. |volume=34 |issue=2 |pages=26–37 |year=2014 |doi=10.1109/MCG.2014.27}}</ref> appear as exceptions, implementing, in different ways, a semantic description of data that can be manipulated in these environments. Avogadro uses the Chemical Markup Language (CML)<ref name="RzepaCML12">{{cite web |url=http://www.xml-cml.org/ |title=Chemical Markup Language |author=Rzepa, H. |publisher=CMLC |date=2012}}</ref> as the format for describing data semantics, and it adds a semantic description layer on top of the data being described. However, the tool leverages neither ontologies nor other knowledge representation formalisms, thus it does not permit reasoning on the described data. | |||
DIVE partially creates ontologies and datasets derived from the input data upon loading. Pre-formatted input in a row/column representation are converted into a SQL-like structure where rows are individuals and columns properties. This data representation conforms to a common data model that the software libraries use. Therefore, creation of links between data values and concepts are possible, and different DIVE components for data presentation (analyses, 3D visualization, etc.) as well as links and relationships between dataset elements can be queried. In addition, DIVE includes a powerful and generic ontology creator directly depending on the type of the input data. However, reasoning on ontologies in DIVE is limited to inheritance between classes. Consequently, only a few ontological relationships are available: is-a, contains, is-part-of, and bound-by. There is no notion of cardinality or logical operators to define the concept classes. Then, it is not possible, for instance, to force the presence of a property, or to impose that only a fixed number of values are associated to a specific property (e.g., a molecule must have at least one atom, an Alanine side-chain has a minimum of three atoms and a maximum of four atoms, etc.). These limitations render the DIVE environment insufficient to solve the problem stated in this paper. | |||
==Using a semantic representation to efficiently store, query, and link heterogeneous structural biology data== | |||
Several important choices have been made to integrate the different technologies required for the establishment of a platform that would allow a proper 3D immersion of users together with an accurate and intelligent way to interact with their data. Our platform heavily relies on the ontology/knowledge base couple. The way to represent and access the data present in the databases is of a crucial importance, and this point led us to ask ourselves the question of the most appropriate formalism for the data representation. | |||
===Knowledge formalism choice=== | |||
The formalism of knowledge representation used in our approach must address the following three rules to properly fit our platform needs: | |||
# Hierarchical data representation via concepts and properties | |||
# Advanced reasoning possibility in order to extend the ontology or the dataset ruled by the ontology | |||
# Efficient query time on the data to stay within interaction time | |||
We mentioned previously that several formalisms exist to create ontologies and define databases. A quick comparison of these formalisms, complementary to their introduction in the previous section, can be found in Table 1. | |||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| class="wikitable" border="1" cellpadding="5" cellspacing="0" width="80%" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" colspan="6"|'''Table 1.''' Comparison of different knowledge representation formalisms with respect to key criteria | |||
|- | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Formalism | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Domain description | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Reasoning on knowledge | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Big data management | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Efficient | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Implementation flexibility | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Conceptual graphs | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|X | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|X | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|X | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|- | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Semantic networks | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|X | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|X | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|X | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|- | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Classical logics | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|X | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|X | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|X | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|X | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|- | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Description logics | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|X | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|X | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|X | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|X | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|- | |||
|- | |||
|} | |||
|} | |||
Our first implementation of a semantic representation of knowledge in molecular biology was applied through conceptual graphs (CG) within Cogitant’s software.<ref name="HuangTheSPHINX93">{{cite journal |title=The SPHINX-II speech recognition system: An overview |journal=Computer Speech & Language |author=Huang, X.; Alleva, F.; Hsiao-Wuen, H. et al. |volume=7 |issue=2 |pages=137–148 |year=1993 |doi=10.1006/csla.1993.1007}}</ref> The use of CGs through the Cogitant API quickly proved to be incompatible with the constraints of the interactive context. This limitation had already been highlighted by the work of Yannick Dennemont<ref name="GenestAPlat98">{{cite journal |title=A platform allowing typed nested graphs: How CoGITo became CoGITaNT |journal=Proceedings from the 1998 International Conference on Conceptual Structures |author=Genest, D.; Salvat, E. |pages=1154–61 |year=1998 |doi=10.1007/BFb0054912}}</ref> with the Prolog CG API, limitations confirmed by our own experience with the Cogitant library in C++. The need for high performance imposed by the interactive context has led us to the path of description logic and semantic web for the representation of knowledge and the efficient extraction of information within a massive fact base to support Visual Analytics functionalities in molecular biology. | |||
==References== | ==References== | ||
| Line 110: | Line 170: | ||
==Notes== | ==Notes== | ||
This presentation is faithful to the original, with only a few minor changes to presentation. Some grammar and punctuation was cleaned up to improve readability. In some cases important information was missing from the references, and that information was added. Nothing else was changed in accordance with the NoDerivatives portion of the license. | This presentation is faithful to the original, with only a few minor changes to presentation. Some grammar and punctuation was cleaned up to improve readability. In some cases important information was missing from the references, and that information was added. The original references after 27 were slightly out of order in the original; due to the way this wiki works, references are listed in the order they appear. Nothing else was changed in accordance with the NoDerivatives portion of the license. | ||
<!--Place all category tags here--> | <!--Place all category tags here--> | ||
Revision as of 02:19, 6 March 2019
| Full article title | Semantics for an integrative and immersive pipeline combining visualization and analysis of molecular data |
|---|---|
| Journal | Journal of Integrative Bioinformatics |
| Author(s) | Trellet, Mikael; Férey, Nicolas; Flotyński, Jakub; Baaden, Marc; Bourdot, Patrick |
| Author affiliation(s) | Bijvoet Center for Biomolecular Research, Université Paris Sud, Poznań Univ. of Economics and Business, Laboratoire de Biochimie Théorique |
| Primary contact | Email: m dot e dot trellet at uu dot nl |
| Year published | 2018 |
| Volume and issue | 15(2) |
| Page(s) | 20180004 |
| DOI | 10.1515/jib-2018-0004 |
| ISSN | 1613-4516 |
| Distribution license | Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International |
| Website | https://www.degruyter.com/view/j/jib.2018.15.issue-2/jib-2018-0004/jib-2018-0004.xml |
| Download | https://www.degruyter.com/downloadpdf/j/jib.2018.15.issue-2/jib-2018-0004/jib-2018-0004.xml (PDF) |
Abstract
The advances made in recent years in the field of structural biology significantly increased the throughput and complexity of data that scientists have to deal with. Combining and analyzing such heterogeneous amounts of data became a crucial time consumer in the daily tasks of scientists. However, only few efforts have been made to offer scientists an alternative to the standard compartmentalized tools they use to explore their data and that involve a regular back and forth between them. We propose here an integrated pipeline especially designed for immersive environments, promoting direct interactions on semantically linked 2D and 3D heterogeneous data, displayed in a common working space. The creation of a semantic definition describing the content and the context of a molecular scene leads to the creation of an intelligent system where data are (1) combined through pre-existing or inferred links present in our hierarchical definition of the concepts, (2) enriched with suitable and adaptive analyses proposed to the user with respect to the current task and (3) interactively presented in a unique working environment to be explored.
Keywords: virtual reality, semantics for interaction, structural biology
Introduction
Recent years have seen a profound change in the way structural biologists interact with their data. New techniques that try to capture the structure and dynamics of bio-molecules have reached an extraordinary high throughput of structural data.[1][2] Scientists must try to combine and analyze data flows from different sources to draw their hypotheses and conclusions. However, despite this increasing complexity, they tend to rely mainly on compartmentalized tools to only visualize or analyze limited portions of their data. This situation leads to a constant back and forth between the different tools and their associated environments. Consequently, a significant amount of time is dedicated to the transformation of data to account for the heterogeneous input data types each tool is allowing.
The need for platforms capable of handling the intricate data flow is then strong. In structural biology, the numerical simulation process is now able to deal with very large and heterogeneous molecular structures. These molecular assemblies may be composed of several million particles and consist of many different types of molecules, including a biologically realistic environment. This overall complexity raises the need to go beyond common visualization solutions and move towards integrated exploration systems where visualization and analysis can be merged.
Immersive environments play an important role in this context, providing both a better comprehension of the three-dimensional structure of molecules, and offering new interaction techniques to reduce the number of data manipulations executed by the experts (see Figure 1). A few studies took advantage of recent developments in virtual reality to enhance some structural biology tasks. Visualization is the first and most obvious task that was improved through new adaptive stereoscopic screens and immersive environments, plunging experts into the very center of their molecules.[3][4][5][6][7] Structure manipulations during specific docking experiments have been improved thanks to the use of haptic devices and audio feedback to drive a simulation.[8] However, if 3D objects can rather easily be represented and manipulated in such environments, the integration of analytical values (energies, distance to reference, etc.)—2D by nature—leads to a certain complexity and is not a solved problem yet. As a consequence, no specific development has been made to set up an immersive platform where the expert could manipulate data coming from different sources to accelerate and improve the development of new hypotheses.
|
This lack of development can also be partly explained by the significant differences between the data handled by the 3D visualization software packages and the analytical tools. On one side, 3D visualization solutions such as PyMol[9], VMD[10], and UnityMol[11] explore and manipulate 3D structure coordinates composing the molecular complex that will be displayed. The scene seen by the user is composed of 3D objects reporting the overall shape of a particular molecule and its environment at a particular state. This scene is static if we are interested in only one state of a given molecule, but is often dynamic when a whole simulated trajectory of conformational changes over time is considered. Analysis tools, on the other side, handle raw numbers, vectors, and matrices in various formats and dimensions, from various input sources depending on the analysis pipeline used to generate them. Their outputs are graphical representations of trends or comparisons between parameters or properties in 1 to N dimensions formatted in a way that experts can quickly understand and use such information to guide their hypotheses.
Some of the aforementioned software do provide tools to gather analyses as static plots aside the 3D visualization space. Interactivity is limited and flexibility mainly depends on the user capability to create and tune scripts to improve the information displayed. We believe that a major improvement of tools available today would bring into play a scenario where the 3D visualization of a molecular event is coupled to monitoring the evolution of analytical properties, e.g., sub-elements such as distance variations and progression of simulation parameters, into a single working environment. The expert would be able to see any action performed in one space (either 3D visualization or analysis) with a coherent graphical impact on the second space to filter or highlight the parameter or sub-ensemble of objects targeted by the expert.
We have developed a pipeline that aims to bring within the same immersive environment the visualization and analysis of heterogeneous data coming from molecular simulations. This pipeline addresses the lack of integrated tools efficiently combining the stereoscopic visualization of 3D objects and the representation/interaction with their associated physicochemical and geometric properties (both 2D and 3D) generated by standard analysis tools and that are either combined to the 3D objects (shape, colour, etc.) or displayed on a dedicated space integrated in the working environment (second mobile screen, 2D integration in the virtual scene, etc.).
In this pipeline, we systematically combine structural and analytical data by using a semantic definition of the content (scientific data) and the context (immersive environments and interfaces). Such a high-level definition can be translated into an ontology from which instances or individuals of ontological concepts can then be created from real data to build a database of linked data for a defined phenomenon. On top of the data collection, an extensive list of possible interactions and actions defined in the ontology and based on the provided data can be computed and presented to the user.
The creation of a semantic definition describing the content and the context of a molecular scene in immersion leads to the creation of an intelligent system where data and 3D molecular representations are (1) combined through pre-existing or inferred links present in our hierarchical definition of the concepts, (2) enriched with suitable and adaptive analyses proposed to the user with respect to the current task, and (3) manipulated by direct interaction allowing to both perform 3D visualization and exploration as well as analysis in a unique immersive environment.
Our method narrows the need for complex interactions by considering what actions the user can perform with the data he is currently manipulating and the means of interaction his immersive environment provides.
We will highlight our developments and the first outcomes of our work through three main sections: the first section attempts to provide a complete background of the usage of semantics in the fields of VR/AR systems and structural biology. In the second section we will describe and justify our implementation choices and how we linked the different technologies highlighted in the previous section. Finally, in a third section, we will show several applications of our platform and its capabilities to address the issues raised previously.
Related works
We present here the state of the art in the two fields related to this paper: the semantic formalism chosen to represent the data and how semantic representations are applied in bioinformatics.
Semantic modeling formalism and semantic web
From classical logic to description logic, from which was derived the "conceptual graph" representation introduced by Sowa[12], many semantic formalisms were used to embed knowledge into applications in order to query and perform reasoning about them.
The conceptual graph formalism represents concepts and properties such as connected graphs and allows complex operations on them. However, it quickly reaches some limitations in terms of performances and implementation flexibility. Classical logic is another well-known formalism but is not broadly used in biology and suffers a lack of implementation tools and libraries. A semantic network limits itself to the representation of concepts and their relations through directed or undirected graphs. It is lacking the possibility to reason over the concepts and their links, reasoning that our intended platform needs. The different requirements of our platform, coupled with our aim to make it as generic as possible, made us choose to use description logics as a formalism for knowledge representation and more precisely the semantic web as underlying standard for the creation of our ontology and the associated knowledge base.
The semantic web has been created by the World Wide Web Consortium under the lead of Tim Berners-Lee, with the aim to share semantic data on the web.[13] It is broadly used by the biggest web companies to uniformly store and share data. It belongs to the family of description logics that use the notions of concepts, roles, and individuals. The concepts are represented by the sub-ensemble of elements in a specific universe, the roles are the links between the elements, and the individuals are the elements of the universe. Each layer of the semantic web (ontology, experimental data, querying process, etc.) has been associated to a language or a format.
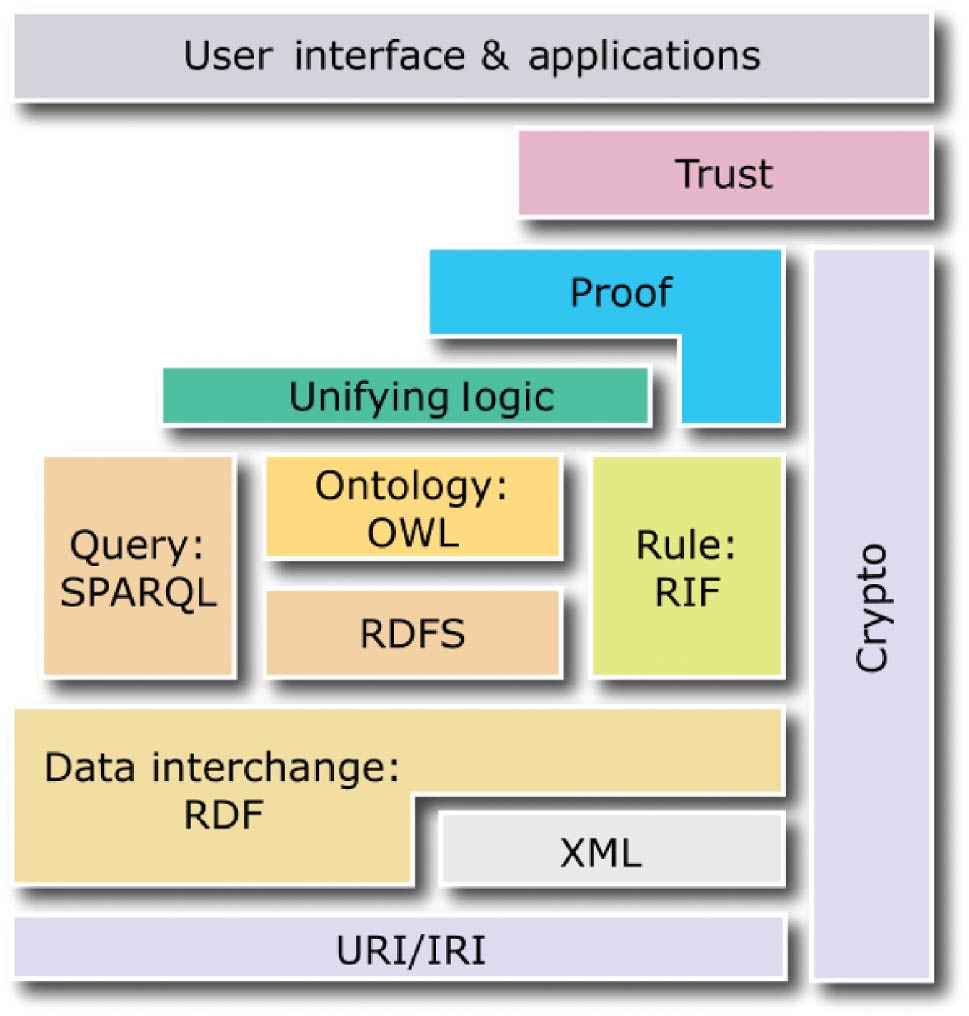
The following four standards create the core of the semantic web and act as the layers evoked previously: the Resource Description Framework (RDF)[14], the Resource Description Framework Schema (RDFS)[15], the Web Ontology Language (OWL)[16], and SPARQL.[17] Whereas the first three standards enable semantic descriptions of data in the form of ontologies and knowledge bases, the last standard enables queries to ontologies and knowledge bases (see Figure 2).
|
RDF is a data model, which allows the creation of statements to describe resources. Each statement is a triple comprised of: a subject (resource described by the statement), a predicate (property of the subject), and an object (literal value or resource identified by a URI, which describes the subject). An example of a triple is: <#Molecule, #has-charge, -1>
RDFS and OWL are semantic web standards that extend the expressiveness of RDF by providing additional concepts. RDFS provides hierarchies of classes and properties as well as property domains and ranges. OWL, built upon RDF and RDFS, provides symmetry, transitivity, equivalence, and restrictions of properties as well as operations on sets of resources. In turn, SPARQL is a query language for ontologies and knowledge bases built using RDF, RDFS, and OWL. Conceptually, in terms of possible operations on data, SPARQL is similar to SQL, as it enables data selection, insertion, update, and removal.
In the semantic web, two types of statements are distinguished. Terminological statements (T-Box) specify conceptualization, classes and properties of resources[18], without describing any particular resources. Assertion statements (A-Box) specify utilization, particular resources (also called individuals or objects), which are instances of classes described by properties with particular values assigned. For example, a T-Box specifies different classes of molecules (different chemical compounds) and properties that can be used to describe them (e.g., charge and the number of neutrons), while an A-Box specifies particular molecules (instances of the classes) with given charges. In this paper, an ontology is a T-Box, while a knowledge base is the union of a T-Box and an A-Box. Ontologies and knowledge bases constitute the foundation of the semantic web across diverse domains and applications. In particular, ontologies can specify schemes of molecular descriptions, while knowledge bases—particular descriptions (instances of such schemes) with individual objects—are used for analysis and visualization. Due to the use of the standards encoded in XML or equivalent formats, ontologies and knowledge bases are interpretable to software, making them intelligible to users. Moreover, since RDFS and OWL are built upon description logics, which are formal knowledge representation techniques, ontologies and knowledge bases can be subject to reasoning, which is a process of inferring implicit (tacit) properties of resources (which have not been explicitly specified by the author) on the basis of their explicitly specified properties.
For instance, from the following triples explicitly specified by the content author:
<my:is-composed-of> <my:is-a> <owl:TransitiveProperty>
<my:Protein> <my:is-composed-of> <my:Amino-acid>
<my:Amino-acid> <my:is-composed-of> <my:Atom>
the following statement can be inferred by software:
<my:Protein> <my:is-composed-of> <my:Atom>
Here, thanks to the definition of property “is-composed-of” as transitive, we can infer that atoms, that compose amino acids, compose as well a protein since amino acids compose proteins. The second statement does not need to be added to the ontology since automatically inferred. This reduces significantly the number of statements to store in the database and potentially allows for more complex inferences.
Ontologies in bioinformatics
On the application side, the use of ontologies in order to standardize knowledge in scientific fields underwent an important and spontaneous growth at the end of the 1990s.[19] Bioinformatics, tightly anchored in structural biology, has used ontologies for a long time. The most significant example is the fast-growing genomic field, in which it became impossible to handle data flow without a proper and standardized organization of the data.[20] The tool Gene Ontology[21] regroups genomic data into a uniform format and a knowledge base. Currently, it is one of the most referred to ontologies in the literature. Rabattu et al.[22] propose an approach to spatio-temporal reasoning on semantic descriptions of an evolving human embryo. Several biological databases or organizations such as UniProtKB1 and the Open Biomedical Ontologies[23] provide ways to access data or ontologies under RDF or OWL format to allow their use in expert tools or specific pipelines. One can also note the open-source project Bio2RDF[24] that aims to build and provide the largest network of "Linked Data for the Life Sciences" using semantic web approaches.
Only a few expert software packages based on ontologies have been developed for structural biology. Avogadro[25] and DIVE[26] appear as exceptions, implementing, in different ways, a semantic description of data that can be manipulated in these environments. Avogadro uses the Chemical Markup Language (CML)[27] as the format for describing data semantics, and it adds a semantic description layer on top of the data being described. However, the tool leverages neither ontologies nor other knowledge representation formalisms, thus it does not permit reasoning on the described data.
DIVE partially creates ontologies and datasets derived from the input data upon loading. Pre-formatted input in a row/column representation are converted into a SQL-like structure where rows are individuals and columns properties. This data representation conforms to a common data model that the software libraries use. Therefore, creation of links between data values and concepts are possible, and different DIVE components for data presentation (analyses, 3D visualization, etc.) as well as links and relationships between dataset elements can be queried. In addition, DIVE includes a powerful and generic ontology creator directly depending on the type of the input data. However, reasoning on ontologies in DIVE is limited to inheritance between classes. Consequently, only a few ontological relationships are available: is-a, contains, is-part-of, and bound-by. There is no notion of cardinality or logical operators to define the concept classes. Then, it is not possible, for instance, to force the presence of a property, or to impose that only a fixed number of values are associated to a specific property (e.g., a molecule must have at least one atom, an Alanine side-chain has a minimum of three atoms and a maximum of four atoms, etc.). These limitations render the DIVE environment insufficient to solve the problem stated in this paper.
Using a semantic representation to efficiently store, query, and link heterogeneous structural biology data
Several important choices have been made to integrate the different technologies required for the establishment of a platform that would allow a proper 3D immersion of users together with an accurate and intelligent way to interact with their data. Our platform heavily relies on the ontology/knowledge base couple. The way to represent and access the data present in the databases is of a crucial importance, and this point led us to ask ourselves the question of the most appropriate formalism for the data representation.
Knowledge formalism choice
The formalism of knowledge representation used in our approach must address the following three rules to properly fit our platform needs:
- Hierarchical data representation via concepts and properties
- Advanced reasoning possibility in order to extend the ontology or the dataset ruled by the ontology
- Efficient query time on the data to stay within interaction time
We mentioned previously that several formalisms exist to create ontologies and define databases. A quick comparison of these formalisms, complementary to their introduction in the previous section, can be found in Table 1.
| ||||||||||||||||||||||||||||||||||||
Our first implementation of a semantic representation of knowledge in molecular biology was applied through conceptual graphs (CG) within Cogitant’s software.[28] The use of CGs through the Cogitant API quickly proved to be incompatible with the constraints of the interactive context. This limitation had already been highlighted by the work of Yannick Dennemont[29] with the Prolog CG API, limitations confirmed by our own experience with the Cogitant library in C++. The need for high performance imposed by the interactive context has led us to the path of description logic and semantic web for the representation of knowledge and the efficient extraction of information within a massive fact base to support Visual Analytics functionalities in molecular biology.
References
- ↑ Zhao, G.; Perilla, J.R.; Yufenyuy, E.L. et al. (2013). "Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics". Nature 497 (7451): 643–6. doi:10.1038/nature12162. PMC PMC3729984. PMID 23719463. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3729984.
- ↑ Zhang, J.; Ma, J.; Liu, D. et al. (2017). "Structure of phycobilisome from the red alga Griffithsia pacifica". Nature 551 (7678): 57–63. doi:10.1038/nature24278. PMID 29045394.
- ↑ van Dam, A.; Forsberg, A.S.; Laidlaw, D.H. et al. (2000). "Immersive VR for scientific visualization: A progress report". IEEE Computer Graphics and Applications 20 (6): 26–52. doi:10.1109/38.888006.
- ↑ Stone. J.E.; Kohlmeyer, A.; Vandivort, K.L.; Schulten, K. (2010). "Immersive molecular visualization and interactive modeling with commodity hardware". Proceedings of the 6th International Conference on Advances in Visual Computing: 382–93. doi:10.1007/978-3-642-17274-8_38.
- ↑ O'Donoghue, S.I.; Goodsell, D.S.; Frangakis, A.S. et al. (2010). "Visualization of macromolecular structures". Nature Methods 7 (3 Suppl.): S42–55. doi:10.1038/nmeth.1427. PMID 20195256.
- ↑ Hirst, J.D.; Glowacki, D.R.; Baaden, M. et al. (2014). "Molecular simulations and visualization: Introduction and overview". Faraday Discussions 169: 9–22. doi:10.1039/c4fd90024c. PMID 25285906.
- ↑ Goddard, T.D., Huang, C.C.; Meng, E.C. et al. (2018). "UCSF ChimeraX: Meeting modern challenges in visualization and analysis". Protein Science 27 (1): 14–25. doi:10.1002/pro.3235. PMC PMC5734306. PMID 28710774. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5734306.
- ↑ Férey, N.; Nelson, J.; Martin, C. et al. (2009). "Multisensory VR interaction for protein-docking in the CoRSAIRe project". Virtual Reality 13: 273. doi:10.1007/s10055-009-0136-z.
- ↑ DeLano, W. (4 September 2000). "The PyMOL Molecular Graphics System". http://pymol.sourceforge.net/overview/index.htm.
- ↑ Humphrey, W.; Dalke, A.; Schulten, K. et al. (1996). "VMD: Visual molecular dynamics". Journal of Molecular Graphics 14 (1): 33–8. doi:10.1016/0263-7855(96)00018-5.
- ↑ Lv, Z.; Tek, A.; Da Silva, F. et al. (2013). "Game on, science - How video game technology may help biologists tackle visualization challenges". PLoS One 8 (3): e57990. doi:10.1371/journal.pone.0057990. PMC PMC3590297. PMID 23483961. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3590297.
- ↑ Sowa, J.F. (1984). Conceptual structures: Information processing in mind and machine. Addison-Wesley Longman Publishing Co. ISBN 0201144727.
- ↑ Berners-Lee, T.; Hendler, J.; Lassila, O. (2001). "The Semantic Web". Scientific American 284: 28–37.
- ↑ Cyganiak, R.; Wood, D.; Lanthaler, M., ed. (25 February 2014). "RDF 1.1 Concepts and Abstract Syntax". World Wide Web Consortium. https://www.w3.org/TR/rdf11-concepts/.
- ↑ Brickley, D.; Guha, R.V., ed. (25 February 2014). "RDF Schema 1.1". World Wide Web Consortium. https://www.w3.org/TR/rdf-schema/.
- ↑ Motik, B.; Patel-Schneider, P.F.; Parsia, B., ed. (11 December 2012). "OWL 2 Web Ontology Language". World Wide Web Consortium. https://www.w3.org/TR/owl2-syntax/.
- ↑ Harris, S.; Seaborne, A., ed. (21 March 2013). "SPARQL 1.1 Query Language". World Wide Web Consortium. https://www.w3.org/TR/sparql11-query/.
- ↑ De Giacomo, G.; Lenzerini, M. (1996). "TBox and ABox Reasoning in Expressive Description Logics". Proceedings of the Fifth International Conference on Principles of Knowledge Representation and Reasoning: 316–27. ISBN 1558604219.
- ↑ Schulze-Kremer, S. (2002). "Ontologies for molecular biology and bioinformatics". In Silico Biology 2 (3): 179–93. PMID 12542404.
- ↑ Schuurman, N.; Leszcynski, A. (2008). "Ontologies for bioinformatics". Bioinformatics and Biology Insights 2: 187—200. PMC PMC2735951. PMID 19812775. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2735951.
- ↑ The Gene Ontology Consortium, Ashburner, M.; Ball, C.A. et al. (2000). "Gene ontology: Tool for the unification of biology". Nature Genetics 25 (1): 25–9. doi:10.1038/75556. PMC PMC3037419. PMID 10802651. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3037419.
- ↑ Rabattu, P.Y.; Massé, B.; Ulliana, F. et al. (2015). "My Corporis Fabrica Embryo: An ontology-based 3D spatio-temporal modeling of human embryo development". Journal of Biomedical Semantics 6: 36. doi:10.1186/s13326-015-0034-0. PMC PMC4582726. PMID 26413258. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4582726.
- ↑ Smith, B.; Ashburner, M.; Rosse, C. et al. (2007). "The OBO Foundry: Coordinated evolution of ontologies to support biomedical data integration". Nature Biotechnology 25 (11): 1251–5. doi:10.1038/nbt1346. PMC PMC2814061. PMID 17989687. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2814061.
- ↑ Belleau, F.; Nolin, M.A;. Tourigny, N. et al. (2008). "Bio2RDF: towards a mashup to build bioinformatics knowledge systems". Journal of Biomedical Informatics 41 (5): 706–16. doi:10.1016/j.jbi.2008.03.004. PMID 18472304.
- ↑ Hanwell, M.D.; Curtis, D.E.; Lonie, D.C. et al. (2012). "Avogadro: An advanced semantic chemical editor, visualization, and analysis platform". Journal of Cheminformatics 4 (1): 17. doi:10.1186/1758-2946-4-17. PMC PMC3542060. PMID 22889332. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3542060.
- ↑ Rysavy, S.J.; Bromley, D.; Daggett, V. (2014). "DIVE: A Graph-Based Visual-Analytics Framework for Big Data". IEEE Computer Graphics and Applications 34 (2): 26–37. doi:10.1109/MCG.2014.27.
- ↑ Rzepa, H. (2012). "Chemical Markup Language". CMLC. http://www.xml-cml.org/.
- ↑ Huang, X.; Alleva, F.; Hsiao-Wuen, H. et al. (1993). "The SPHINX-II speech recognition system: An overview". Computer Speech & Language 7 (2): 137–148. doi:10.1006/csla.1993.1007.
- ↑ Genest, D.; Salvat, E. (1998). "A platform allowing typed nested graphs: How CoGITo became CoGITaNT". Proceedings from the 1998 International Conference on Conceptual Structures: 1154–61. doi:10.1007/BFb0054912.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. Some grammar and punctuation was cleaned up to improve readability. In some cases important information was missing from the references, and that information was added. The original references after 27 were slightly out of order in the original; due to the way this wiki works, references are listed in the order they appear. Nothing else was changed in accordance with the NoDerivatives portion of the license.