Difference between revisions of "Journal:Terminology spectrum analysis of natural-language chemical documents: Term-like phrases retrieval routine"
Shawndouglas (talk | contribs) (Added content. Saving and adding more.) |
Shawndouglas (talk | contribs) (Added content. Saving and adding more.) |
||
| Line 152: | Line 152: | ||
====Recognition of tokens with special meaning==== | ====Recognition of tokens with special meaning==== | ||
The significant part of text pre-processing stage is selection of individual tokens being the words of general English and recognition of various meaningful text strings which are: the general scientific terms (actually performed at the final terminology spectrum building stage but described here for convenience); tokens denoting chemical elements, stable isotopes and measurement units; tokens which cannot be a part of any terms in any way. This part of work is performed using specially developed dictionaries described in details in Table 1. | The significant part of text pre-processing stage is selection of individual tokens being the words of general English and recognition of various meaningful text strings which are: the general scientific terms (actually performed at the final terminology spectrum building stage but described here for convenience); tokens denoting chemical elements, stable isotopes and measurement units; tokens which cannot be a part of any terms in any way. This part of work is performed using specially developed dictionaries described in details in Table 1. | ||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| class="wikitable" border="1" cellpadding="5" cellspacing="0" width="60%" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" colspan="4"|'''Table 1.''' Developed/modified dictionaries used for recognition of general English words, general chemical science terms and tokens with special meaning | |||
|- | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Dictionary/''Usage for'' | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Description | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Reference | |||
! style="background-color:#e2e2e2; padding-left:10px; padding-right:10px;"|Examples | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|General chemical science terms<br /> <br />''Selection of general terms (chemical and from related fields of physics, mathematics …)'' | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|~7500 General scientific terms in chemistry, physics and mathematics<br /> <br />IUPAC Compendium is used | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|http://goldbook.iupac.org/<br /> <br />IUPAC Compendium of Chemical Terminology (''Gold Book'') | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Naphthenes, solvation energy, osmotic pressure, reaction dynamics … | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|General English words dictionary<br /> <br />''Selection of general English wordsGeneral chemical science terms'' | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|~58,000 general English words. It is based on Corncob Lowercase Dictionary modified by us for stated goals. 566 words were excluded, which are often used in scientific terminology | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Modified Corncob Lowercase list of more than 57,000 English words http://ru.scribd.com/doc/147594864/<br /> <br />Corncob Lowercase (see Additional file 3 for excluded words) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Abbreviate, academic, accelerate …<br /> <br />'''Excluded''': Abrasion, absorption, aerosol … | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Stop list<br /> <br />''Filtering tokens which are not part of terms in any way'' | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|~2060 tokens. List contains the words, abbreviations and so on, which cannot be incorporated into any term-like phrases | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Proprietary design (see Additional file 4) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|e.g., de, ca., fig., al., co-exist, et, etc., i.e., ltd … | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Stable isotopes<br /> <br />''Filtering n-grams containing digits'' | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|~250 isotopes. It is based on The Berkeley Laboratory Isotopes Project’s isotopes database | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Proprietary design, based on The Berkeley Laboratory Isotopes Project’s DB: http://ie.lbl.gov/education/isotopes.htm (see Additional file 5) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|1H, 2H, 3He, 4He, 6Li, 7Li … | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Chemical elements signs<br /> <br />''Filtering n-grams containing digits'' | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|~126 chemical elements. It is based on periodic table | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Proprietary design, based on periodic table (see Additional file 6) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|H, He, Li, Be, B, C, N, O, F … | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Measurement units<br /> <br />''Filtering n-grams containing units of measure'' | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|~100 records now, partially based on IUPAC ''Gold Book'' | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|Proprietary design, partially based on http://goldbook.iupac.org/ (see Additional file 7) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"|(a.u.), (ev), a.u, °C, ppm, kV, mol, g<sup>−1</sup>, ml<sup>−1</sup>, gcat, gcat h … | |||
|- | |||
|} | |||
|} | |||
Some extra explanation needs to be given on the general English dictionary, the stop list dictionary and the procedure of recognition of general scientific terms. | |||
More than 560 words either found in scientific terminology (for instance: "acid", "alcohol", "aldehyde", "alloy", "aniline", etc.) or occurring in composite terms (for example, "abundant" may be part of the term "most abundant reactive intermediates") were excluded from the original version of the Corncob Lowercase Dictionary. | |||
The ''IUPAC Compendium of Chemical Terminology'' (the only well-known and time-proven dictionary) is used as a source of general chemistry terms. To find the best way to match an n-gram to a scientific term from the compendium, a number of experiments have been performed which resulted in the following criteria: | |||
1. N-gram is considered a general scientific term if all n-gram tokens are the words of a certain IUPAC ''Gold Book'' term, regardless of their order; and | |||
2. If (n − 1) of n-gram tokens coincide with the (n − 1) words of an IUPAC ''Gold Book'' term, and the remaining word is among other terms in the dictionary, then the n-gram is considered a general scientific term too. | |||
Some examples may be given. The n-gram "RADIAL CONCENTRATION GRADIENT" is a general scientific term because the phrase "concentration gradient" is in the compendium and the word "radial" is part of the term "radial development." The n-gram "CONTENT CATALYTIC ACTIVITY" is a general term because the term "catalytic activity content" is present in the compendium and differs from the n-gram only by word order. The n-gram "TOLUENE ADSORPTION CAPACITY" is not considered a general term, despite the fact that two words coincide with the term "absorption capacity," because the remaining word "TOLUENE" is special and is not found in the compendium. The n-gram "COBALT ACETATE DECOMPOSITION" is not considered a general term either as only the term "decomposition" may be found. | |||
The final comment is about the stop list dictionary that, at first glance, may look like a set of arbitrary words. But, actually, it is based on a series of observations performed with the set of wrongly identified term-like phrases by the earlier version of the terminology analysis system. | |||
====Strict filtering==== | |||
The last step in the text pre-processing stage is strict filtering developed to remove unnecessary words and meaningless combinations of symbols. If at least one n-gram token is labeled by the strict filtering tag ("rubbish" : "true") then such an n-gram is not considered a term-like phrase. At this stage, certain character sequences — as described by the filtering rules (Table 2) and not exempt by the list of exceptions (Table 3) — are looked for. They are successive digits, special symbols, measurement units, symbols of chemical elements, brackets and so on. Custom regular expressions and standard dictionaries described in Table 1 are used for this procedure. A general scheme of strict filtering parsing is illustrated in Fig. 4. | |||
Revision as of 20:23, 6 June 2016
| Full article title | Terminology spectrum analysis of natural-language chemical documents: Term-like phrases retrieval routine |
|---|---|
| Journal | Journal of Cheminformatics |
| Author(s) | Alperin, Boris L.; Kuzmin, Andrey O.; Ilina, Ludmila Y.; Gusev, Vladimir D.; Salomatina, Natalia V.; Parmon, Valentin, N. |
| Author affiliation(s) | Boreskov Institute of Catalysis, Sobolev Institute of Mathematics, Novosibirsk State University |
| Primary contact | Email: kuzmin [at] catalysis.ru |
| Year published | 2016 |
| Volume and issue | 8 |
| Page(s) | 22 |
| DOI | 10.1186/s13321-016-0136-4 |
| ISSN | 1758-2946 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | http://jcheminf.springeropen.com/articles/10.1186/s13321-016-0136-4 |
| Download | http://jcheminf.springeropen.com/track/pdf/10.1186/s13321-016-0136-4 (PDF) |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
Background: This study seeks to develop, test and assess a methodology for automatic extraction of a complete set of ‘term-like phrases’ and to create a terminology spectrum from a collection of natural language PDF documents in the field of chemistry. The definition of ‘term-like phrases’ is one or more consecutive words and/or alphanumeric string combinations with unchanged spelling which convey specific scientific meanings. A terminology spectrum for a natural language document is an indexed list of tagged entities including: recognized general scientific concepts, terms linked to existing thesauri, names of chemical substances/reactions and term-like phrases. The retrieval routine is based on n-gram textual analysis with a sequential execution of various ‘accept and reject’ rules with taking into account the morphological and structural information.
Results: The assessment of the retrieval process, expressed quantitatively with a precision (P), recall (R) and F1-measure, which are calculated manually from a limited set of documents (the full set of text abstracts belonging to five EuropaCat events were processed) by professional chemical scientists, has proved the effectiveness of the developed approach. The term-like phrase parsing efficiency is quantified with precision (P = 0.53), recall (R = 0.71) and F1-measure (F1 = 0.61) values.
Conclusion: The paper suggests using such terminology spectra to perform various types of textual analysis across document collections. This sort of terminology spectrum may be successfully employed for text information retrieval, for reference database development, to analyze research trends in subject fields of research and to look for the similarity between documents.
|
|
Keywords: Terminology spectrum, natural language text analysis, n-Gram analysis, term-like phrases retrieval, text information retrieval
Background
The current situation in chemistry, as in any other field of natural science, can be characterized by a substantial growth of texts in natural languages (research papers, conference proceedings, patents, etc.), still being the most important sources of scientific knowledge and experimental data, information about modern research trends and terminology used in the subject areas of science. It greatly increases the value of such powerful information systems as Scopus®, SciFinder®, and Reaxys® which are capable of handling large text document databases and especially those fitted with advanced text information retrieval capabilities. In fact, both efficiency and productivity of modern scientific research in chemistry depend rigorously on quality and completeness of its information support, which is oriented firstly on advanced and flexible reference search, discovering and analysing of text information to afford the most relevant answers to user questions (substances, reactions, relevant patents or journal articles). The main ideas and developments in the information retrieval methods coupled with techniques of full text analysis are now well described and examined.[1]
In conventional information systems, the majority of text information retrieval and discovery methods are based on using specific sets of pre-defined document metadata, e.g. keywords or indexes of terms characterizing the texts content. User queries are converted using an index into information requests expressed by a combination of Boolean terms while bringing into play the vector space and terms weight. Probabilistic approaches may also be employed to take into account such features as terms distribution, co-occurrence information and their relationships derived from information retrieval thesauri (IRT) to include them into analytic process. Any kind of such indexes have to primarily be produced and updated manually by trained experts, but now the possibilities of automated index development attracts closer attention.
It is assumed that the structural foundation of any scientific text is its terminology, which may be represented, in principle, by advanced IRT. However, it leads to difficulties in applying conventional IRTs in practical information text analysis procedures because of limitations inherent in them. Typically, such thesauri are made manually in a very labor-intensive process and often are constructed to reflect the general terminology only. Terms from thesauri originally represent a formally written description of scientific conceptions and definitions which may not exactly match the real usage and spelling used in scientific texts. Moreover, a thesaurus developed for one type of text may be less efficient or not applicable when used with another. A good example is the IUPAC Gold Book compendium of chemical nomenclature, terminology, units and definition recommendations.[2] Terminology drafted by experts of IUPAC spans a wide range of chemistry but does not describe any field in detail and represents only a well-established upper level of scientific terminology. Summarizing, IRT based text analysis alone is unable to solve the problem of the variability of scientific texts written in natural languages because the accuracy of matching thesaurus terms with real text phrases leaves much to be desired.
It should also be noted that the language of science is evolving faster than that of natural language, especially in chemistry and molecular biology. Thus, the analysis of terminology of subject text collection should be done automatically using both primitive extraction and sophisticated knowledge-based parsing. Only automated data analysis can process and reveal the variety of term-like word combinations in the constantly changing world of scientific publications. Automated parsing and analysis of document collections or isolated documents for term-like phrases can also help to discover various contexts in which the same scientific terminology is used in different publications or even parts of the same publication.
There is nothing new in the idea of automated term retrieval. Typically, the terminology analysis of text content is focused on recognition of chemical entities and automatic keyphrase extraction aimed to provide a limited set of keywords which might characterize and classify the document as a whole. Two main strategies are usually applied: machine self-learning and usage of various dictionaries with automated selection rules (heuristics) coupled with calculated features[3], such as TF-IDF.[4][5] Therefore, keyphrase retrieval procedures typically involve the following stages: initial text pre-processing; selecting a candidate to a keyphrase; applying rules to each candidate; and compiling a list of keyphrases.[6] A few existing systems had been analyzed in terms of precision (P), recall (R) and F1-score attainable for existing keyphrase extraction datasets. For such well-known systems as Wingnus, Sztergak, and KP-Mminer, these values are reported as P = 0.34÷0.40, R = 0.11÷0.14, and F1 = 0.17÷0.20.[6] Open-Source Chemistry Analysis Routines (OSCAR4)[7] and ChemicalTagger[8] NLP may also be mentioned as tools for the recognition of named chemical entities and for parsing and tagging the language of text publications in chemistry.
However, there are some inherent shortcomings in the above mentioned keyphrase extraction approaches due to the presence of a significant amount of cases where a limited set of automatically selected top ranked keyphrases does not properly describe the document in details (e.g., a paper may contain the description of a specific procedure of catalyst preparation while not being the main subject of the paper). It may also be seen from the aforementioned values of P, R and F that in many cases the extracted keyphrases do not match the keyphrases selected by experts to an adequate degree. Exact matching of keyphrases is a rather rare event, partially due to the difficulties of taking into account nearly similar phrases, for instance, semantically similar phrases. On the other hand, even though the widely used n-gram analysis can build a full spectrum of token sequences present in the text, it may also produce a great level of noise, making it difficult to use them. Some attempts have been made to take into account the semantic similarity of n-grams and to differentiate between rubbish and candidates to plausible keyphrases.[9][10]
The problem of automatic recognition of scientific terms in natural language texts has been explored in recent decades.[11] That research has shown that taking into account the linguistic information may improve the terms extraction efficiency. The information about grammatical structure of multi-word scientific terms, their text variants, and the context of their usage may be represented as a set of lexico-syntactic patterns. For instance, values of P, R and F-measure equal to 73.1, 53.6 and 61.8 percent respectively for term extraction from scientific texts (only in Russian) on computer science and physics were obtained.[12]
A "terminology spectrum" of a natural language publication may be defined as an indexed list of tagged token sequences with calculated weights, such as recognized general scientific notions, terms linked to existing thesauri, names of chemical entities and "term-like phrases." The term-like phrases are not exactly the keyphrases or terms in the usual sense (like published in thesauri). Such term-like phrases are defined here as one or more consecutive tokens (represented by words and/or alphanumeric strings combinations), which convey specific scientific meaning with unchanged spelling and context as in a real text document. For instance, a term-like phrase may look similar to a specific generally used term but with different spelling or word order reflecting the usage of the term in a different context in natural language environment. Consequently, they may describe real text content and the essence of real processes that the scientific research handles, which makes the analysis of such phrases extremely useful. That sort of terminology spectrum of a natural language publication may be considered as some kind of knowledge representation of a text and may be successfully employed in various information retrieval strategies, text analysis and reference systems.[13]
The present work is aimed to develop and test the methodology of automated retrieval of full terminology spectrum from any natural language chemical text collections in PDF format, with term-like phrases selection being the central part of the procedure. The retrieval routine is based on n-gram text analysis with sequential execution of a complex grouping of "accept" and ‘"eject" rules while taking into account the morphological and structural information. The term "n-gram" denotes here a text string or a sequence of n consecutive words or tokens presented in a text. Numerical assessment of automated term-like phrases retrieval process efficiency done in the paper is calculated by comparing automatically extracted term-like phrases and those manually selected by experts.
Methods
Text collection used for experiments
Chemical catalysis is a foundation of chemical industry and represents a very complex field of scientific and technological research. It includes chemistry, various subject fields of physics, chemical engineering, material science and a lot more. One of the most representative research conferences in catalysis is the European Congress on Catalysis or EuropaCat, which has been chosen as a source of scientific texts covering the wide range of themes of research. A set of abstracts of EuropaCat conferences of 2013, 2011, 2009, 2007, and 2005 (about 6000 documents from all five Congress events) has been used for textual analysis in the present study. All abstracts are in PDF format.
General description of terminology spectrum retrieval process
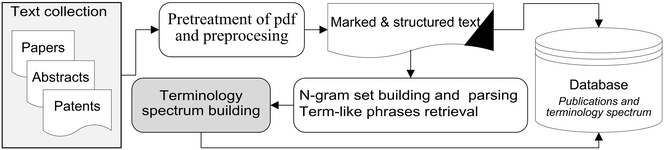
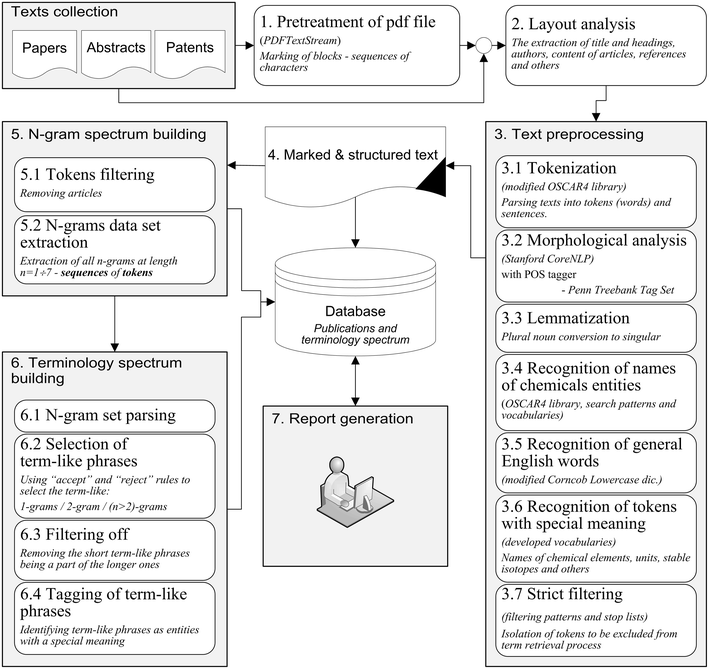
The developed system of terminology spectrum analysis consists of the following sequentially running procedures or steps, as depicted in Fig. 1.
|
The server side of the terminology spectrum analysis system runs on Java SE 6 platform and the client is a PHP web application to view texts and the results of terminology analysis. To store all data collected in the terminology retrieval process, the cross-platform document-oriented database MongoDB is used.[14] The choice in favor of MongoDB was conditioned by the need to process nested n-gram structures up to level seven.
The main stages and analytic methods involved in the process are discussed in the following sections.
Text materials conversion with PdfTextStream library
The scientific texts are mainly published in PDF format which does not typically contain any information about document structure and therefore is not suitable for immediate text analysis. Thus, at first, a document has to be preprocessed by converting a PDF file into text format and analyzing its structure (highlighting titles, authors, headings, references, etc.) with the aim to make the text suitable for further content information retrieval (see Fig. 2). The following steps are used with PdfTextStream library[15] (stages 1–2 on Fig. 1) to make such a PDF transformation (for a detailed example see Additional File 1):
|
1. Isolate text blocks which have the same formatting (e.g. bold, underline and etc.).
2. Remove empty blocks and merge blocks located on the same text row.
3. Analyze the document structure by classifying each block as containing information about the publication title, the headings, authors, organizations, e-mails, references and content. To perform such analysis a set of special taggers has been developed which are executed sequentially to analyze and tag each text block. Taggers utilize such features as the position of the first and last rows of text block, its text formatting, the position of a block of text on a page, etc. All developed taggers have been adjusted to handle each conference event individually.
4. Filter text blocks to remove unclassified text blocks, for instance, situated before the publication title, because such blocks typically contain useless and already known information about a conference or journal.
5. Unify special symbols (such as variants of the dash, hyphen, and quote characters), removal of space characters placed before brackets in writings of crystal indexes, etc. Regular expressions are used.
Text pre-processing
The text pre-processing stage (step three in Fig. 1) is used to transform a text document obtained from stages one and two into a unified structured format with markup. During this stage the text is split into individual words and sentences (tokenization) followed by a morphological analysis that includes: highlighting objects such as formulas and chemical entities, removing unnecessary words and meaningless combinations of symbols, and recognizing general English words and tokens with special meaning (units, stable isotopes, acronyms, etc.). The result of this stage is a fully marked structured text to be stored in the database. The following steps are involved in the text pre-processing stage.
Tokenization
A tokenizer from the OSCAR4 library is used for splitting a text into words, phrases and other meaningful elements. The tokenizer has been adapted for better handling of chemical texts.
The present study established that the original OSCAR4 tokenizer, in view of our needs, had some shortcomings. The first issue was a separation of tokens with a hyphen "-", which often led to mistakes in recognizing compound terms. To overcome this issue, the parts of the source code which are responsible for splitting tokens with hyphens were commented out (see Additional File 2). Next was a problem where some complex tokens, representing various chemical compositions, were considered by the tokenizer as a sequence of tokens (see Fig. 3). In such cases it was necessary to combine those isolated tokens into an integrated one. The modified tokenizing procedure now makes merging of tandem tokens separated with either the "/" or ":" characters, provided that they are marked by OSCAR4 tag CM or incorporate a chemical element symbol sign. Additionally, tokens that look as "number %" and are situated at the beginning of such a phrase describing chemical compositions are merged into the integral token too (see Fig. 3).
https://static-content.springer.com/image/art%3A10.1186%2Fs13321-016-0136-4/MediaObjects/13321_2016_136_Fig3_HTML.gif
|
An example of the work of the modified tokenizer is shown on Fig. 3. Blue frames hold the tokens identified by modified OSCAR4 tokenizer. Additional red frames outline tokens which are combined into integral ones. Such tokens are marked with the isolated tag COMP. This tag is used by accept rule ChemUnigramRule to identify one-word n-grams describing chemical compositions.
Then the position of a token in the text is determined. Splitting the series of tokens into sentences finalizes the tokenization process, which is realized with the help of the WordToSentenceAnnotator routine of Stanford CoreNLP library.[16][17]
Morphological analysis and labeling tokens with their POS tags
Morphological analysis (Stanford CoreNLP library[18] is used) maps each word with a set of part-of-speech tags (Penn Treebank Tag Set[19] by Stanford CoreNLP is used). Typical tags used in the research are: NN (plural NNS) — noun; VB — verb; JJ — adjective; CD — ordinal numeral, etc. For the full information about the POS tags used by terminology spectrum building procedure, see Table 4 (later in the paper).
Lemmatization
Lemmatization is the process of grouping together different inflected word forms so they can be treated as a single item. But, in the present work, lemmatization is only used to replace nouns in the plural form with their lemmas. Preliminary experiments demonstrate that additional lemmatization is not helpful and leads to a significant loss of meaningful information (for example, reforming process leads to reform and process lemmas with the loss of the name of a very important modern industrial chemical process in refining).
Recognition of names of chemical entities
Meta-information about names of chemical entities is very important in various term-like phrases retrieval strategies. The open source OSCAR4 (Open Source Chemistry Analysis Routines)[7][20] software package is applied for selection and semantic annotation of chemical entities across a text. Among a variety of tags and attributes utilized by OSCAR4 routine only the following ones are used in the present study:
1. CM — chemical term (chemical name, formula or acronym);
2. RN — reaction (for example, epoxidation, dehydrogenation, hydrolysis, etc.);
3. ONT — ontology term (for example, glass, adsorption, cation, etc.).
When a token is a part of some recognized chemical entity the token gets the same OSCAR4 tag as a whole entity.
Recognition of tokens with special meaning
The significant part of text pre-processing stage is selection of individual tokens being the words of general English and recognition of various meaningful text strings which are: the general scientific terms (actually performed at the final terminology spectrum building stage but described here for convenience); tokens denoting chemical elements, stable isotopes and measurement units; tokens which cannot be a part of any terms in any way. This part of work is performed using specially developed dictionaries described in details in Table 1.
| ||||||||||||||||||||||||||||||||
Some extra explanation needs to be given on the general English dictionary, the stop list dictionary and the procedure of recognition of general scientific terms.
More than 560 words either found in scientific terminology (for instance: "acid", "alcohol", "aldehyde", "alloy", "aniline", etc.) or occurring in composite terms (for example, "abundant" may be part of the term "most abundant reactive intermediates") were excluded from the original version of the Corncob Lowercase Dictionary.
The IUPAC Compendium of Chemical Terminology (the only well-known and time-proven dictionary) is used as a source of general chemistry terms. To find the best way to match an n-gram to a scientific term from the compendium, a number of experiments have been performed which resulted in the following criteria:
1. N-gram is considered a general scientific term if all n-gram tokens are the words of a certain IUPAC Gold Book term, regardless of their order; and
2. If (n − 1) of n-gram tokens coincide with the (n − 1) words of an IUPAC Gold Book term, and the remaining word is among other terms in the dictionary, then the n-gram is considered a general scientific term too.
Some examples may be given. The n-gram "RADIAL CONCENTRATION GRADIENT" is a general scientific term because the phrase "concentration gradient" is in the compendium and the word "radial" is part of the term "radial development." The n-gram "CONTENT CATALYTIC ACTIVITY" is a general term because the term "catalytic activity content" is present in the compendium and differs from the n-gram only by word order. The n-gram "TOLUENE ADSORPTION CAPACITY" is not considered a general term, despite the fact that two words coincide with the term "absorption capacity," because the remaining word "TOLUENE" is special and is not found in the compendium. The n-gram "COBALT ACETATE DECOMPOSITION" is not considered a general term either as only the term "decomposition" may be found.
The final comment is about the stop list dictionary that, at first glance, may look like a set of arbitrary words. But, actually, it is based on a series of observations performed with the set of wrongly identified term-like phrases by the earlier version of the terminology analysis system.
Strict filtering
The last step in the text pre-processing stage is strict filtering developed to remove unnecessary words and meaningless combinations of symbols. If at least one n-gram token is labeled by the strict filtering tag ("rubbish" : "true") then such an n-gram is not considered a term-like phrase. At this stage, certain character sequences — as described by the filtering rules (Table 2) and not exempt by the list of exceptions (Table 3) — are looked for. They are successive digits, special symbols, measurement units, symbols of chemical elements, brackets and so on. Custom regular expressions and standard dictionaries described in Table 1 are used for this procedure. A general scheme of strict filtering parsing is illustrated in Fig. 4.
References
- ↑ Salton, G. (1991). "Developments in Automatic Text Retrieval". pp. 974–980. doi:10.1126/science.253.5023.974. PMID 17775340.
- ↑ "IUPAC Gold Book". International Union of Pure and Applied Chemistry. 2014. http://goldbook.iupac.org/.
- ↑ Hussey, R.; Williams, S.; Mitchell, R. (2012). "Automatic keyphrase extraction: A comparison of methods". eKNOW, Proceedings of The Fourth International Conference on Information Process, and Knowledge Management: 18–23. ISBN 9781612081816.
- ↑ Eltyeb, S.; Salim, N. (2014). "Chemical named entities recognition: a review on approaches and applications". Journal of Cheminformatics 6: 17. doi:10.1186/1758-2946-6-17. PMC PMC4022577. PMID 24834132. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4022577.
- ↑ Gurulingappa, H.; Mudi, A.; Toldo, L.; Hofmann-Apitus, M.; Bhate, J. (2013). "Challenges in mining the literature for chemical information". RSC Advances 2013 (3): 16194-16211. doi:10.1039/C3RA40787J.
- ↑ 6.0 6.1 Kim, S.N.; Madelyan, O.; Kan, M.-Y.; Baldwin, T. (2013). "Automatic keyphrase extraction from scientific articles". Language Resources and Evaluation 47 (3): 723–742. doi:10.1007/s10579-012-9210-3.
- ↑ 7.0 7.1 Jessop, D.M.; Adams, S.E.; Willighagen, E.L.; Hawizy, L.; Murray-Rust, P. (2011). "OSCAR4: A flexible architecture for chemical text-mining". Journal of Cheminformatics 3: 41. doi:10.1186/1758-2946-3-41. PMC PMC3205045. PMID 21999457. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3205045.
- ↑ Hawizy, L.; Jessop, D.M.; Adams, N.; Murray-Rust, P. (2011). "ChemicalTagger: A tool for semantic text-mining in chemistry". Journal of Cheminformatics 3: 17. doi:10.1186/1758-2946-3-17. PMC PMC3117806. PMID 21575201. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3117806.
- ↑ "Re-examining automatic keyphrase extraction approaches in scientific articles". MWE '09 Proceedings of the Workshop on Multiword Expressions: Identification, Interpretation, Disambiguation and Applications: 9–16. 2009. ISBN 9781932432602.
- ↑ "Approximate matching for evaluating keyphrase extraction". RANLP '09: International Conference on Recent Advances in Natural Language Processing: 484–489. 2009.
- ↑ Castellvi, M.T.C.; Bagot, R.E.; Palatresi, J.V. (2001). "Automatic term detection: A review of current systems". In Bourigault, D.; Jacquemin, C.; L'Homme, M.-C.. Recent Advances in Computational Terminology. John Benjamins Publishing Company. pp. 53–87. doi:10.1075/nlp.2.04cab. ISBN 9789027298164.
- ↑ Bolshakova, E.I.; Efremova, N.E. (2015). "A Heuristic Strategy for Extracting Terms from Scientific Texts". In Khachay, M.Y.; Konstantinova, N.; Panchenko, A.; Ignatov, D.I.; Labunets, V.G.. Analysis of Images, Social Networks and Texts. Springer International Publishing. pp. 297-307. doi:10.1007/978-3-319-26123-2_29. ISBN 9783319261232.
- ↑ Salton, G.; Buckley, C. (1991). "Global Text Matching for Information Retrieval". pp. 1012–1015. doi:10.1126/science.253.5023.1012. PMID 17775345.
- ↑ Chodorow, K.; Dirolf, M. (2010). MongoDB: The Definitive Guide. O'Reilly Media. ISBN 9781449381561.
- ↑ "PDFxStream". Snowtide Informatics Systems, Inc. 2016. https://www.snowtide.com/.
- ↑ "Stanford CoreNLP – A suite of core NLP tools". Github. 2016. http://stanfordnlp.github.io/CoreNLP/.
- ↑ "The Stanford CoreNLP Natural Language Processing Toolkit". Proceedings of the 52nd Annual Meeting of the Association for Computational Linguistics: System Demonstrations: 55–60. 2014. doi:10.3115/v1/P14-5010.
- ↑ Toutanova, K.; Klein, D.; Manning, C.D.; Singer, Y. (2003). "Feature-rich part-of-speech tagging with a cyclic dependency network". NAACL '03 Proceedings of the 2003 Conference of the North American Chapter of the Association for Computational Linguistics on Human Language Technology 1: 173–180. doi:10.3115/1073445.1073478.
- ↑ Taylor, A.; Marcus, M.; Santorini, B. (2003). "The Penn Treebank: An Overview". In Abeillé, A.. Text, Speech and Language Technology. 20. Springer Netherlands. pp. 5–22. doi:10.1007/978-94-010-0201-1_1. ISBN 978-94-010-0201-1.
- ↑ "Semantic enrichment of journal articles using chemical named entity recognition". ACL '07 Proceedings of the 45th Annual Meeting of the ACL on Interactive Poster and Demonstration Sessions: 45–48. 2007.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. Numerous grammar errors were also corrected throughout the entire text.