Journal:The GAAIN Entity Mapper: An active-learning system for medical data mapping
| Full article title | The GAAIN Entity Mapper: An active-learning system for medical data mapping |
|---|---|
| Journal | Frontiers in Neuroinformatics |
| Author(s) | Ashish, N.; Dewan, P.; Toga, A.W. |
| Author affiliation(s) | University of Southern California at Los Angeles |
| Primary contact | Email: nashish@loni.usc.edu |
| Editors | Van Ooyen, A. |
| Year published | 2016 |
| Volume and issue | 9 |
| Page(s) | 30 |
| DOI | 10.3389/fninf.2015.00030 |
| ISSN | 1662-5196 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | http://journal.frontiersin.org/article/10.3389/fninf.2015.00030/full |
| Download | http://journal.frontiersin.org/article/10.3389/fninf.2015.00030/pdf (PDF) |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
This work is focused on mapping biomedical datasets to a common representation, as an integral part of data harmonization for integrated biomedical data access and sharing. We present GEM, an intelligent software assistant for automated data mapping across different datasets or from a dataset to a common data model. The GEM system automates data mapping by providing precise suggestions for data element mappings. It leverages the detailed metadata about elements in associated dataset documentation such as data dictionaries that are typically available with biomedical datasets. It employs unsupervised text mining techniques to determine similarity between data elements and also employs machine-learning classifiers to identify element matches. It further provides an active-learning capability where the process of training the GEM system is optimized. Our experimental evaluations show that the GEM system provides highly accurate data mappings (over 90 percent accuracy) for real datasets of thousands of data elements each, in the Alzheimer's disease research domain. Further, the effort in training the system for new datasets is also optimized. We are currently employing the GEM system to map Alzheimer's disease datasets from around the globe into a common representation, as part of a global Alzheimer's disease integrated data sharing and analysis network called GAAIN. GEM achieves significantly higher data mapping accuracy for biomedical datasets compared to other state-of-the-art tools for database schema matching that have similar functionality. With the use of active-learning capabilities, the user effort in training the system is minimal.
Keywords: data mapping, machine learning, active learning, data harmonization, common data model
Background and significance
This paper describes a software solution for biomedical data harmonization. Our work is in the context of the “GAAIN” project in the domain of Alzheimer's disease data. However, this solution is applicable to any biomedical or clinical data harmonization in general. GAAIN — the Global Alzheimer's Association Interactive Network — is a data sharing federated network of Alzheimer's disease datasets from around the globe. The aim of GAAIN is to create a network of Alzheimer's disease data, researchers, analytical tools and computational resources to better our understanding of this disease. A key capability of this network is also to provide investigators with access to harmonized data across multiple, independently created Alzheimer's datasets.
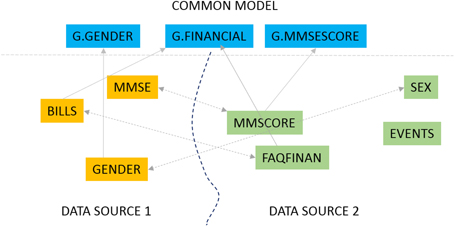
Our primary interest is in biomedical data sharing and specifically harmonized data sharing. Harmonized data from multiple data providers has been curated to a unified representation after reconciling the different formats, representation, and terminology from which it was derived.[1][2] The process of data harmonization can be resource-intensive and time-consuming; the present work describes a software solution to significantly automate that process. Data harmonization is fundamentally about data alignment, the establishment of correspondence between related or identical data elements across different datasets. Consider the very simple example of a data element capturing the gender of a subject that is defined as “SEX” in one dataset, “GENDER” in another and “M/F” in yet another. When harmonizing data, a unified element is needed to capture this gender concept and to link (align) the individual elements in different datasets with this unified element. This unified element is the “G.GENDER” element as illustrated in Figure 1.
|
The data mapping problem can be solved in two ways as illustrated in Figure 1. We could map elements across two datasets, for instance match the element “GENDER” from one data source (DATA SOURCE 1 in Figure 1) to the element “SEX” in a second source (DATA SOURCE 2). We could also map elements from one dataset to elements from a common data model. A common data model is a uniform representation which all data sources or providers in a data sharing network agree to adopt. The fundamental mapping task is the same in both. Also, the task of data alignment is inevitable regardless of the data sharing model one employs. In a centralized data sharing model[3], where we create a single unified store of data from multiple data sources, the data from any data source must be mapped and transformed to the unified representation of the central repository. In federated or mediated approaches to data sharing[1] individual data sources (such as databases) must be mapped to a “global” unified model through mapping rules. The common data model approach, which is also the GAAIN approach, also requires us to map and transform every dataset to the (GAAIN) common data model. This kind of data alignment or mapping can be labor intensive in biomedical and clinical data integration case studies.[4] A single dataset typically has thousands of distinct data elements of which a large subset must be accurately mapped. And it is widely acknowledged that data sharing and integration processes need to be simplified and made less resource-intensive for data sharing, for them to become a viable solution for the medical and clinical data sharing domain as well as the more general enterprise information integration domain.[5] The GEM system is designed to achieve this by providing automated assistance to developers for such data alignment or mapping.
The GEM data mapping approach is centered on exploiting the information in the data documentation, typically in the form of data dictionaries associated with the data. The importance of data dictionary documentation, and for Alzheimer's data in particular, has been articulated by Morris et al.[6] These data dictionaries contain detailed descriptive information and metadata about each data element in the dataset. Our solution is based on extracting this rich metadata in data dictionaries, developing element similarity measures based on text mining of the element descriptions, and employing machine-learning classifiers to meaningfully combine multiple indicative factors for an element match.
Materials and methods
Here we report the second version of the GEM system.[7] The first version (GEM-1.0)[7], deployed in December 2014 is a knowledge driven system. Element matches are determined in a heuristic manner based on element similarity derived off the element metadata in data dictionaries. In this second version (GEM-2.0), completed in April 2015, we added machine-learning based classification to the system. We have further added capabilities of active-learning[8] where the user effort in training the machine-learning classifiers in the system is minimized.
While GEM-2.0 data mapping is powered by machine-learning classification, it employs the element metadata extraction developed in GEM-1.0 for synthesizing features required for classification. In this section we will describe the extraction of metadata from data dictionaries and element similarity metrics as developed in GEM-1.0. We then describe the machine-learning classification capabilities developed in GEM-2.0. The last sub-section describes the active-learning capability for training the system for new datasets, in an efficient manner. Before describing the system however, we clarify some terminology and definitions.
- A dataset is a source of data. For example, a dataset provided by “ADNI” is a source.
- A data dictionary is the document associated with a dataset, which defines the terms used in the dataset.
- A data element is an individual “atomic” unit of information in a dataset, for instance a field or a column in a table in a database or in a spreadsheet.
- The documentation for each data element in a data dictionary is called element metadata or element information.
- A mapping or element mapping is a one-to-one relationship across two data elements, coming from different sources.
- Mappings are created across two distinct sources. The element that we seek to match is called the query element. The source we must find matches from is called the target source, and the source of the query element is called the query source. (Note: A common data model may also be treated as a target source.)
- For any element, the GEM system provides a set of suggestions (typically five). We refer to this set as the window of suggestions.
Metadata extraction for element similarity
Medical datasets, including datasets in domains such as Alzheimer's disease, are typically documented in data dictionaries. The data dictionary provides information about each element, including descriptions of the data element and other details such as its data type, range or set of possible values etc.
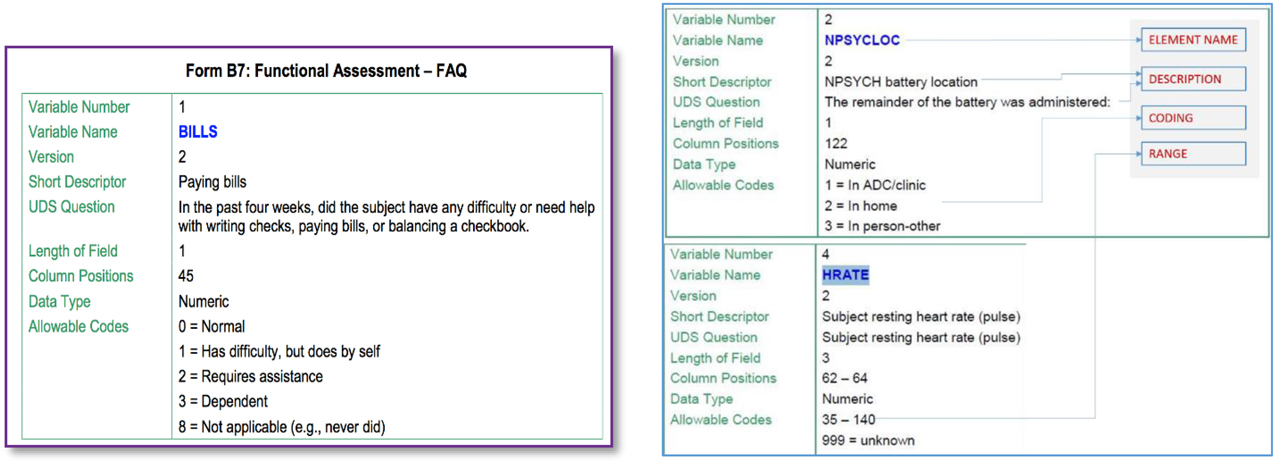
Figure 2 illustrates some snippets from a data dictionary for a particular Alzheimer's disease dataset. The element described (on the left) is called “BILLS” and includes a short as well as a more complete description of this data element. We are also provided information about its type (numeric code in this case) and also the possible values it can take, i.e., one of {0,1,2,3,8}. Such element information is the basis for identifying GEM element mappings.
|
As a data mapping system GEM operates as follows:
- Elements are matched based on a metric of element similarity that is assessed by using the above illustrated metadata for each element.
- Such metadata is first extracted from the data dictionaries (of the datasets to be matched) and stored in a metadata database.
- Elements are matched from a source dataset to a target dataset. For instance we may be interested in finding mappings between two datasets such as ADNI and say “NACC” of which one would be the source and the other the target.
- The target may also be a common data model. The elements of the common model are treated as (data) elements of a target data model.
- In determining the possible matches for a given data element, some of the metadata constraints are used for “blocking,” i.e., elimination from consideration of improbable matches. For instance an element (such as say BILLS) that takes one of five distinct coded values (as illustrated above) cannot match to an element such as BLOOD PRESSURE that takes values in the range 90–140.
- Probable matches for a data element are determined using element similarity. The element similarity is a score that captures how well (or not) the corresponding text descriptions of two data elements match. This text description similarity is computed using two approaches.
- One is to use a TF-IDF cosine distance on a bag-of-words representation of the text descriptions. TF-IDF, short for term frequency–inverse document frequency, is a numerical statistic that is intended to reflect how important a word is to a document in a collection or corpus.[9]
- Another approach requires building a topic model[10] over the element text descriptions and using the topic distribution to derive the similarity between two element text descriptions.
References
- ↑ 1.0 1.1 Doan, A.; Halevy, A.; Ives, Z. (2012). Principles of Data Integration (1st ed.). Elsevier. pp. 520. ISBN 9780123914798.
- ↑ Ohmann, C.; Kuchinke, W. (2009). "Future developments of medical informatics from the viewpoint of networked clinical research: Interoperability and integration". Methods of Information in Medicine 48 (1): 45–54. doi:10.3414/ME9137. PMID 19151883.
- ↑ "National Database for Autism Research". NIH. 2015. https://ndar.nih.gov/.
- ↑ Ashish, N.; Ambite, J.L.; Muslea, M.; Turner, J.A. (2010). "Neuroscience data integration through mediation: An (F)BIRN case study". Frontiers in Neuroinformatics 4: 118. doi:10.3389/fninf.2010.00118. PMC PMC3017358. PMID 21228907. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3017358.
- ↑ Halevy, A.Y.; Ashish, N.; Bitton, D. et al. (2005). "Enterprise information integration: Successes, challenges and controversies". Proceedings of the 2005 ACM SIGMOD International Conference on Management of Data: 778–787. doi:10.1145/1066157.1066246.
- ↑ Morris, J.C.; Weintraub, S.; Chui, H.C. et al.. "The Uniform Data Set (UDS): Clinical and cognitive variables and descriptive data from Alzheimer Disease Centers". Alzheimer Disease and Associated Disorders 20 (4): 210–6. doi:10.1097/01.wad.0000213865.09806.92. PMID 17132964.
- ↑ 7.0 7.1 Ashish, N.; Dewan, P.; Ambite, J.; Toga, A. (2015). "GEM: The GAAIN Entity Mapper". Proceedings of the 11th International Conference on Data Integration in Life Sciences: 13–27. doi:10.1007/978-3-319-21843-4_2. PMC PMC4671774. PMID 26665184. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4671774.
- ↑ Cohn, D.A.; Ghahramani, Z.; Jordan, M.I. (1996). "Active Learning with Statistical Models". Journal of Artificial Intelligence Research 4: 129–145. doi:10.1613/jair.295.
- ↑ Robertson, S. (2004). "Understanding inverse document frequency: On theoretical arguments for IDF". Journal of Documentation 60 (5): 503–520. doi:10.1108/00220410410560582.
- ↑ Blei, D.M. (2012). "Probabilistic topic models". Communications of the ACM 55 (4): 77–84. doi:10.1145/2133806.2133826.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. References are in order of appearance rather than alphabetical order (as the original was). Some grammar, punctuation, and minor wording issues have been corrected.