Difference between revisions of "Journal:The growing need for microservices in bioinformatics"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) (Finished adding rest of content.) |
||
| Line 18: | Line 18: | ||
|website = [http://www.jpathinformatics.org/article.asp?issn=2153-3539;year=2016;volume=7;issue=1;spage=45;epage=45;aulast=Williams http://www.jpathinformatics.org] | |website = [http://www.jpathinformatics.org/article.asp?issn=2153-3539;year=2016;volume=7;issue=1;spage=45;epage=45;aulast=Williams http://www.jpathinformatics.org] | ||
|download = [http://www.jpathinformatics.org/temp/JPatholInform7145-5650543_154145.pdf http://www.jpathinformatics.org/temp/JPatholInform7145-5650543_154145.pdf] (PDF) | |download = [http://www.jpathinformatics.org/temp/JPatholInform7145-5650543_154145.pdf http://www.jpathinformatics.org/temp/JPatholInform7145-5650543_154145.pdf] (PDF) | ||
}} | }} | ||
| Line 59: | Line 54: | ||
The widely adopted framework for microservices bundles are referred to as "containers." A container is essentially an encapsulated and immutable version of an application, coupled with the bare-minimum operating system components required for execution. Libraries for running containers are currently available for Linux and soon will be available for Windows Server 2016.<ref name="FultonTheWin15">{{cite web |url=http://thenewstack.io/the-windows-and-linux-container-era-is-here/ |title=The Windows and Linux Container Era is Here |author=Fulton, S.M. |work=The New Stack |publisher=Alex Williams Media, LLC |date=19 August 2015 |accessdate=05 January 2016}}</ref> Containers utilize a relatively small footprint and require less overhead than virtual machines; running containers requires little more than installing a light-weight program on the host. (Virtual machines require the installation of a complete operating system in addition to the host operating system. This additional operating system sequesters system resources, such as central processing unit cores and random access memory, reducing what is available to the host. In contrast, containers use the same Linux kernel as the host system and share system resources only as needed) The host and container operating systems are generally unaware of each other, allowing for disparate systems to run harmoniously on a common host. This decouples the microservice(s) from the underlying physical infrastructure, allowing development effort to be focused solely on functionality. | The widely adopted framework for microservices bundles are referred to as "containers." A container is essentially an encapsulated and immutable version of an application, coupled with the bare-minimum operating system components required for execution. Libraries for running containers are currently available for Linux and soon will be available for Windows Server 2016.<ref name="FultonTheWin15">{{cite web |url=http://thenewstack.io/the-windows-and-linux-container-era-is-here/ |title=The Windows and Linux Container Era is Here |author=Fulton, S.M. |work=The New Stack |publisher=Alex Williams Media, LLC |date=19 August 2015 |accessdate=05 January 2016}}</ref> Containers utilize a relatively small footprint and require less overhead than virtual machines; running containers requires little more than installing a light-weight program on the host. (Virtual machines require the installation of a complete operating system in addition to the host operating system. This additional operating system sequesters system resources, such as central processing unit cores and random access memory, reducing what is available to the host. In contrast, containers use the same Linux kernel as the host system and share system resources only as needed) The host and container operating systems are generally unaware of each other, allowing for disparate systems to run harmoniously on a common host. This decouples the microservice(s) from the underlying physical infrastructure, allowing development effort to be focused solely on functionality. | ||
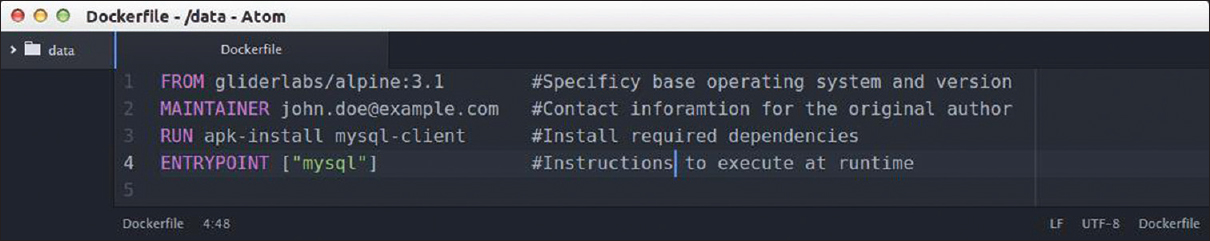
== | Designing a container is a fairly straightforward process. A configuration file [Figure 2] describes the base operating system, build options, dependencies required for the service to run, and optional initialization scripts. This simple text file fully defines the container and can be used to instantiate the container on any capable host without additional setup. In contrast, traditional application deployment requires a specific build for each host, and applications with conflicting dependencies will cause significant problems. | ||
[[File:Fig2 Williams JofPathInformatics2016 7.jpg|800px]] | |||
{{clear}} | |||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| border="0" cellpadding="5" cellspacing="0" width="800px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 2''' Example configuration file for a simple container providing a MySQL client service. Adapted from https://hub.docker.com/r/gliderlabs/alpine</blockquote> | |||
|- | |||
|} | |||
|} | |||
The infrastructure described thus far allows for flexibility in scaling applications, especially when scaling across multiple nodes or even across heterogeneous networks. Well-designed containers are stateless (stateless in the sense that output is determined solely by the current input and independent of previous inputs) or at least having no internal state, meaning they can be aborted and restarted without losing information. This allows for services to be created ''ad hoc'' and then deleted when they are no longer needed. Running services on an as-needed basis allow for optimal use of system resources. Similarly, portability of containers allows for automated and nearly instantaneous scaling of services and resources. When demand fluctuates, it is possible to scale the application up, either on existing hardware or alternatively, by incorporating additional cloud resources. At a functional level, this is accomplished by spawning new containers as needed and then scaling them back when appropriate by destroying dormant containers. This flexibility allows for a measured approach to scaling infrastructure, rather than purchasing fixed quantities of hardware to handle either typical or peak application loads. | |||
Regardless of the benefits and increasing popularity observed in the industry<ref name="DelVecchioDocker15">{{cite web |url=https://www.linkedin.com/pulse/docker-scores-best-ever-net-score-etr-history-thomas-delvecchio |title=Docker Scores the Best Ever NET Score in ETR History |author=DelVecchio, T. |work=LinkedIn Pulse |publisher=LinkedIn Corporation |date=17 April 2015 |accessdate=06 January 2016}}</ref>, the question of whether similar benefits can be reaped by the bioinformatics community by embracing this design practice remains to be answered. We propose it can be a valuable tool and offer benefits beyond what has previously been discussed. As bioinformatics tools grow in complexity, it is difficult for small [[laboratory|laboratories]] to maintain the in-house expertise required to modify and further develop all stages of a complete informatics pipeline. It will, however, become feasible to experiment with algorithms and use a wider variety of tools, as the services are divided into more manageable, interchangeable modules. Containers also facilitate the use of cloud-based platform-as-a-service (PaaS) providers, avoiding the potentially prohibitive costs associated with buying state-of-the-art hardware, which may itself sit idle for the majority of its life as it quickly becomes obsolete. PaaS providers offer access to nearly inexhaustible computing resources that can be retained for the length of an application's execution for a reasonable fee. An application built on containers can be constructed anew with each execution, run on a data set, output the results, and then dismantle itself, thus releasing its resources without manual intervention. Such automation significantly reduces operating costs and overhead of hardware, in addition to reducing the in-house technical expertise required. | |||
The very nature of containers lends itself to supporting collaboration and transparency. Researchers can easily share their work without worrying about replicating a specific development environment or sharing access to computing resources between institutions. Simply sharing the configuration files for each microservice allows others to create an exact replica of the original development environment. This same method can be used to share experimental results in an entirely new way, allowing for entire algorithms and pipelines to be published alongside the dataset. Users can then easily recreate the pipeline, rerun the calculations, and validate the experimental results, and then compare those results with locally derived data run through the same pipeline. The [[National Institutes of Health]] have made their desires known for improved rigor and reproducibility in research.<ref name="NIHRigor16">{{cite web |url=https://grants.nih.gov/reproducibility/index.htm |title=Rigor and Reproducibility |publisher=National Institutes of Health |date=02 November 2015 |accessdate=08 January 2016}}</ref> The methods and practices inherent to microservice architecture would seamlessly address this intent for increased transparency and reproducibility. | |||
Going further, an opportunity exists to leverage distributed source control tools, such as Git or [[Apache Subversion|Subversion]], to create a shared repository of bioinformatics tools and algorithms. Such a service, if embraced by the community, would offer a hub of documented, open-source tools that could easily be shared, annotated, and enhanced by the community. While many open-source tools are currently available, these projects often rely upon specific versions of libraries and therefore may not easily interface with even a minority of other extant tools. Finally, adapting these open-source applications to a different workflow may involve a technical challenge similar in scope to the original development effort. Conversely, a curated hub of bioinformatics tools utilizing the microservices framework, being thoroughly vetted and maintained by the community, would promote reuse of these valuable resources. | |||
==Conclusions== | |||
Developing custom in-house software in support of bioinformatics research is a resource-intensive effort requiring complementary expertise in software development and data science. Those without the requisite resources are generally compelled to rely upon commercially supported software, often at significant expense and with the additional drawbacks of limited flexibility and customization. Utilization of a containerized framework lowers barriers of entry, increases development efficiency, facilitates reuse of proven code modules, and allows for simplified verification of algorithm integrity through transparency. This is accomplished by leveraging freely available open-source tools that have been adopted by leaders in the IT industry. | |||
On a more practical note, while there are multiple implementations of the container methodology in industry, the Docker ecosystem of tools is the most widely publicized, and it can serve as a useful source of information for learning the basics of Linux containers. The concepts found in the core online Docker documentation can be applied broadly, but such information is also helpful in support of bioinformatics projects that might make use of the Docker model. NextFlow<ref name="NFDocker16">{{cite web |url=https://www.nextflow.io/docs/latest/docker.html |title=Docker Containers |publisher=International Cancer Genome Consortium |date=18 November 2015 |accessdate=12 August 2016}}</ref>, the PanCancer Launcher<ref name="ICGCWorking15">{{cite web |url=http://icgc.org/index.php?q=working-pancancer-data-aws |title=Working with PanCancer Data on AWS |work=Nextflow's Documentation |publisher=Nextflow Development Group |date=2016 |accessdate=12 August 2016}}</ref> from the International Cancer Genome Consoritum, and BaseSpace<ref name="IlluminaBaseSpace16">{{cite web |url=https://developer.basespace.illumina.com/docs/content/documentation/native-apps/native-app-overview |title=BaseSpace Native App Engine Overview |work=Documentation |publisher=Illumina, Inc |date=2016 |accessdate=12 August 2016}}</ref> from Illumina, are a few examples of contemporary tools that take advantage of Docker to help manage associated pipelines. There are also at least two projects aimed at sharing bioinformatics based Docker images: bioboxes (http://bioboxes.org) and BioDocker (http://biodocker.org). Both contain explicit examples on how to containerize, locally-developed bioinformatics tools, and associated repositories of available tools supported by the community. Wider adoption of any of the above-enumerated services could be of great value to the research and bioinformatics communities-at-large. | |||
While use-cases supporting discovery are certainly fertile ground for microservices realizing an immediate impact within pathology, there is certainly potential for a much broader impact in the clinical realm as well. Mission-critical services, such as billing and interface engines, could similarly benefit from the high availability capabilities afforded from this approach. In terms of healthcare production IT settings, being able to distribute new communication standards, such as Fast Healthcare Interoperability Resources, by use of microservices could promote adherence to a common standard and facilitate faster adoption by the community. | |||
Large, monolithic, and proprietary applications have been the ''de facto'' standard for pathology and healthcare in general. Indeed, vendors appear to be in a continuing milieu of market forces, where they are actively incentivized to maintain the status quo, since profit margins can be effectively maintained by controlling access to the data. However, that paradigm has been evolving rapidly outside of healthcare and there are even signs that it may be slowly starting to change within healthcare (as well with the adoption of [[health information exchange]]s). By gaining familiarity and expertise with microservice architecture, we believe pathologists can contain ever-expanding IT costs, reduce the likelihood of IT implementation mishaps and failures, and perhaps most importantly, greatly elevate the level of service conferred to our many types of customers. | |||
==Financial support and sponsorship== | |||
Nil. | |||
==Conflicts of interest== | ==Conflicts of interest== | ||
There are no conflicts of interest. | |||
==References== | ==References== | ||
Latest revision as of 22:19, 20 December 2016
| Full article title | The growing need for microservices in bioinformatics |
|---|---|
| Journal | Journal of Pathology Informatics |
| Author(s) | Williams, Christopher L.; Sica, Jeffrey C.; Killen, Robert T.; Balis, Ulysses G.J. |
| Author affiliation(s) | University of Michigan |
| Primary contact | Email: Available w/ login |
| Year published | 2016 |
| Volume and issue | 7 |
| Page(s) | 45 |
| DOI | 10.4103/2153-3539.194835 |
| ISSN | 2153-3539 |
| Distribution license | Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported |
| Website | http://www.jpathinformatics.org |
| Download | http://www.jpathinformatics.org/temp/JPatholInform7145-5650543_154145.pdf (PDF) |
Abstract
Objective: Within the information technology (IT) industry, best practices and standards are constantly evolving and being refined. In contrast, computer technology utilized within the healthcare industry often evolves at a glacial pace, with reduced opportunities for justified innovation. Although the use of timely technology refreshes within an enterprise's overall technology stack can be costly, thoughtful adoption of select technologies with a demonstrated return on investment can be very effective in increasing productivity and at the same time, reducing the burden of maintenance often associated with older and legacy systems.
In this brief technical communication, we introduce the concept of microservices as applied to the ecosystem of data analysis pipelines. Microservice architecture is a framework for dividing complex systems into easily managed parts. Each individual service is limited in functional scope, thereby conferring a higher measure of functional isolation and reliability to the collective solution. Moreover, maintenance challenges are greatly simplified by virtue of the reduced architectural complexity of each constitutive module. This fact notwithstanding, rendered overall solutions utilizing a microservices-based approach provide equal or greater levels of functionality as compared to conventional programming approaches. Bioinformatics, with its ever-increasing demand for performance and new testing algorithms, is the perfect use-case for such a solution. Moreover, if promulgated within the greater development community as an open-source solution, such an approach holds potential to be transformative to current bioinformatics software development.
Context: Bioinformatics relies on nimble IT framework which can adapt to changing requirements.
Aims: To present a well-established software design and deployment strategy as a solution for current challenges within bioinformatics
Conclusions: Use of the microservices framework is an effective methodology for the fabrication and implementation of reliable and innovative software, made possible in a highly collaborative setting.
Keywords: Bioinformatics, crowd sourcing, defect analysis, failure mode analysis, information technology, microservices, pathology, reliability engineering, software engineering
Introduction
The modern software ecosystem is rapidly changing. The emergence of cloud computing has spurred developers to compartmentalize applications into small, easily maintainable functional units that can be replicated on demand. This application segmentation is commonly referred to as microservices (or microservice architecture). The enabling technology for microservices was first introduced into the UNIX kernel as early as 1979[1], but this field has only recently experienced an exponential growth in popularity. Many industry leaders, dependent on their information technology (IT) infrastructure, have embraced this model, including Amazon, Capital One Financial Corp., Google, and PayPal Holdings Inc.[2], among many others.
Discussion
Microservice architecture has taken hold in application design for a number of reasons [Figure 1], but ultimately, the fundamental driver has been the opportunity for significant increase in productivity. Gains are achieved by dividing larger, more complex applications into small, manageable projects that encapsulate common functionality. Each microservice represents a "black-box" with a well-defined application programming interface (API) bundled with its own integral resources, thus eliminating external dependencies. This encapsulation simplifies the division of labor for teams of software developers and reduces the inherent overhead of multiple developers working on the same sections of code. The narrow scope of functionality of a single microservice allows for thorough testing and optimization of each component in isolation, leading to higher reliability and performance of the overall parent application. The encapsulated nature of the microservice methodology inherently allows for future reuse of existing modules with minimal effort. A portfolio of proven, production-use microservices can be valuable for accelerating the development of subsequent projects.
|
This architecture can also allow for the improvement of application stability and security. It does so by eliminating interdependency of components, as well as handling service failures gracefully, without the need to halt an entire application. Where high-availability application architectures are required, a system health check can be run as a background task, querying the various components for their status. When a service is malfunctioning, a "watch-dog" routine can shut down the service in question and then restart it. Generally, this process can take place without degrading the performance of the application as a whole. A somewhat infamous example occurred at Netflix prior to their transition to a distributed, microservices architecture.[7] Apparently, a semicolon in a segment of code (or rather, the lack of a semicolon) took their service offline for an extended period of time, while engineers from across the enterprise attempted to track down the source of the disruption. Security can be enforced through carefully-crafted APIs. A breach of any particular module does not expose the entirety of the application, only the limited information accessed by the particular service. By severely restricting data flow between public-facing services and elements handling sensitive data, the risk posed by intrusion can be mitigated. This segregation of services also allows for real-time security patching of individual modules as vulnerabilities are exposed, which is a large improvement over the historical method of coordinating planned downtime to patch the various vulnerable components.
The widely adopted framework for microservices bundles are referred to as "containers." A container is essentially an encapsulated and immutable version of an application, coupled with the bare-minimum operating system components required for execution. Libraries for running containers are currently available for Linux and soon will be available for Windows Server 2016.[8] Containers utilize a relatively small footprint and require less overhead than virtual machines; running containers requires little more than installing a light-weight program on the host. (Virtual machines require the installation of a complete operating system in addition to the host operating system. This additional operating system sequesters system resources, such as central processing unit cores and random access memory, reducing what is available to the host. In contrast, containers use the same Linux kernel as the host system and share system resources only as needed) The host and container operating systems are generally unaware of each other, allowing for disparate systems to run harmoniously on a common host. This decouples the microservice(s) from the underlying physical infrastructure, allowing development effort to be focused solely on functionality.
Designing a container is a fairly straightforward process. A configuration file [Figure 2] describes the base operating system, build options, dependencies required for the service to run, and optional initialization scripts. This simple text file fully defines the container and can be used to instantiate the container on any capable host without additional setup. In contrast, traditional application deployment requires a specific build for each host, and applications with conflicting dependencies will cause significant problems.
|
The infrastructure described thus far allows for flexibility in scaling applications, especially when scaling across multiple nodes or even across heterogeneous networks. Well-designed containers are stateless (stateless in the sense that output is determined solely by the current input and independent of previous inputs) or at least having no internal state, meaning they can be aborted and restarted without losing information. This allows for services to be created ad hoc and then deleted when they are no longer needed. Running services on an as-needed basis allow for optimal use of system resources. Similarly, portability of containers allows for automated and nearly instantaneous scaling of services and resources. When demand fluctuates, it is possible to scale the application up, either on existing hardware or alternatively, by incorporating additional cloud resources. At a functional level, this is accomplished by spawning new containers as needed and then scaling them back when appropriate by destroying dormant containers. This flexibility allows for a measured approach to scaling infrastructure, rather than purchasing fixed quantities of hardware to handle either typical or peak application loads.
Regardless of the benefits and increasing popularity observed in the industry[9], the question of whether similar benefits can be reaped by the bioinformatics community by embracing this design practice remains to be answered. We propose it can be a valuable tool and offer benefits beyond what has previously been discussed. As bioinformatics tools grow in complexity, it is difficult for small laboratories to maintain the in-house expertise required to modify and further develop all stages of a complete informatics pipeline. It will, however, become feasible to experiment with algorithms and use a wider variety of tools, as the services are divided into more manageable, interchangeable modules. Containers also facilitate the use of cloud-based platform-as-a-service (PaaS) providers, avoiding the potentially prohibitive costs associated with buying state-of-the-art hardware, which may itself sit idle for the majority of its life as it quickly becomes obsolete. PaaS providers offer access to nearly inexhaustible computing resources that can be retained for the length of an application's execution for a reasonable fee. An application built on containers can be constructed anew with each execution, run on a data set, output the results, and then dismantle itself, thus releasing its resources without manual intervention. Such automation significantly reduces operating costs and overhead of hardware, in addition to reducing the in-house technical expertise required.
The very nature of containers lends itself to supporting collaboration and transparency. Researchers can easily share their work without worrying about replicating a specific development environment or sharing access to computing resources between institutions. Simply sharing the configuration files for each microservice allows others to create an exact replica of the original development environment. This same method can be used to share experimental results in an entirely new way, allowing for entire algorithms and pipelines to be published alongside the dataset. Users can then easily recreate the pipeline, rerun the calculations, and validate the experimental results, and then compare those results with locally derived data run through the same pipeline. The National Institutes of Health have made their desires known for improved rigor and reproducibility in research.[10] The methods and practices inherent to microservice architecture would seamlessly address this intent for increased transparency and reproducibility.
Going further, an opportunity exists to leverage distributed source control tools, such as Git or Subversion, to create a shared repository of bioinformatics tools and algorithms. Such a service, if embraced by the community, would offer a hub of documented, open-source tools that could easily be shared, annotated, and enhanced by the community. While many open-source tools are currently available, these projects often rely upon specific versions of libraries and therefore may not easily interface with even a minority of other extant tools. Finally, adapting these open-source applications to a different workflow may involve a technical challenge similar in scope to the original development effort. Conversely, a curated hub of bioinformatics tools utilizing the microservices framework, being thoroughly vetted and maintained by the community, would promote reuse of these valuable resources.
Conclusions
Developing custom in-house software in support of bioinformatics research is a resource-intensive effort requiring complementary expertise in software development and data science. Those without the requisite resources are generally compelled to rely upon commercially supported software, often at significant expense and with the additional drawbacks of limited flexibility and customization. Utilization of a containerized framework lowers barriers of entry, increases development efficiency, facilitates reuse of proven code modules, and allows for simplified verification of algorithm integrity through transparency. This is accomplished by leveraging freely available open-source tools that have been adopted by leaders in the IT industry.
On a more practical note, while there are multiple implementations of the container methodology in industry, the Docker ecosystem of tools is the most widely publicized, and it can serve as a useful source of information for learning the basics of Linux containers. The concepts found in the core online Docker documentation can be applied broadly, but such information is also helpful in support of bioinformatics projects that might make use of the Docker model. NextFlow[11], the PanCancer Launcher[12] from the International Cancer Genome Consoritum, and BaseSpace[13] from Illumina, are a few examples of contemporary tools that take advantage of Docker to help manage associated pipelines. There are also at least two projects aimed at sharing bioinformatics based Docker images: bioboxes (http://bioboxes.org) and BioDocker (http://biodocker.org). Both contain explicit examples on how to containerize, locally-developed bioinformatics tools, and associated repositories of available tools supported by the community. Wider adoption of any of the above-enumerated services could be of great value to the research and bioinformatics communities-at-large.
While use-cases supporting discovery are certainly fertile ground for microservices realizing an immediate impact within pathology, there is certainly potential for a much broader impact in the clinical realm as well. Mission-critical services, such as billing and interface engines, could similarly benefit from the high availability capabilities afforded from this approach. In terms of healthcare production IT settings, being able to distribute new communication standards, such as Fast Healthcare Interoperability Resources, by use of microservices could promote adherence to a common standard and facilitate faster adoption by the community.
Large, monolithic, and proprietary applications have been the de facto standard for pathology and healthcare in general. Indeed, vendors appear to be in a continuing milieu of market forces, where they are actively incentivized to maintain the status quo, since profit margins can be effectively maintained by controlling access to the data. However, that paradigm has been evolving rapidly outside of healthcare and there are even signs that it may be slowly starting to change within healthcare (as well with the adoption of health information exchanges). By gaining familiarity and expertise with microservice architecture, we believe pathologists can contain ever-expanding IT costs, reduce the likelihood of IT implementation mishaps and failures, and perhaps most importantly, greatly elevate the level of service conferred to our many types of customers.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- ↑ Topjian, J. (13 November 2013). "Contain your enthusiasm - Part One: A history of operating system containers". Tech Radar Blog. Cybera. http://www.cybera.ca/news-and-events/tech-radar/contain-your-enthusiasm-part-one-a-history-of-operating-system-containers/. Retrieved 05 January 2016.
- ↑ 2.0 2.1 Miller, M. (5 October 2015). "Innovate or Die: The Rise of Microservices". The Wall Street Journal. Dow Jones & Company, Inc. http://blogs.wsj.com/cio/2015/10/05/innovate-or-die-the-rise-of-microservices/. Retrieved 05 January 2016.
- ↑ "GE Uses Docker to Enable Self-Service for their Developers". Docker, Inc. 2016. https://www.docker.com/customers/ge-uses-docker-enable-self-service-their-developers. Retrieved 29 August 2016.
- ↑ "Flexible, Portable, Secure". Hewlett Packard Enterprise Development LP. 2016. https://www.docker.com/customers/ge-uses-docker-enable-self-service-their-developers. Retrieved 28 August 2016.
- ↑ Ho, A. (11 November 2015). "Microservices at Amazon". Developer Blog. Apigee, Inc. https://apigee.com/about/blog/developer/microservices-amazon. Retrieved 28 August 2016.
- ↑ Piesche, S. (23 March 2015). "Microservices at Constant Contact". Tech Blog. Constant Contact, Inc. https://apigee.com/about/blog/developer/microservices-amazon. Retrieved 28 August 2016.
- ↑ "Why You Can’t Talk About Microservices Without Mentioning Netflix". SmartBear Software. 8 December 2015. http://blog.smartbear.com/microservices/why-you-cant-talk-about-microservices-without-mentioning-netflix/. Retrieved 08 august 2016.
- ↑ Fulton, S.M. (19 August 2015). "The Windows and Linux Container Era is Here". The New Stack. Alex Williams Media, LLC. http://thenewstack.io/the-windows-and-linux-container-era-is-here/. Retrieved 05 January 2016.
- ↑ DelVecchio, T. (17 April 2015). "Docker Scores the Best Ever NET Score in ETR History". LinkedIn Pulse. LinkedIn Corporation. https://www.linkedin.com/pulse/docker-scores-best-ever-net-score-etr-history-thomas-delvecchio. Retrieved 06 January 2016.
- ↑ "Rigor and Reproducibility". National Institutes of Health. 2 November 2015. https://grants.nih.gov/reproducibility/index.htm. Retrieved 08 January 2016.
- ↑ "Docker Containers". International Cancer Genome Consortium. 18 November 2015. https://www.nextflow.io/docs/latest/docker.html. Retrieved 12 August 2016.
- ↑ "Working with PanCancer Data on AWS". Nextflow's Documentation. Nextflow Development Group. 2016. http://icgc.org/index.php?q=working-pancancer-data-aws. Retrieved 12 August 2016.
- ↑ "BaseSpace Native App Engine Overview". Documentation. Illumina, Inc. 2016. https://developer.basespace.illumina.com/docs/content/documentation/native-apps/native-app-overview. Retrieved 12 August 2016.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added.