Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Fix) |
Shawndouglas (talk | contribs) (Updated article of the week text) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File: | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig6 Ogle FrontBigData2021 4.jpg|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:Named data networking for genomics data management and integrated workflows|Named data networking for genomics data management and integrated workflows]]"''' | ||
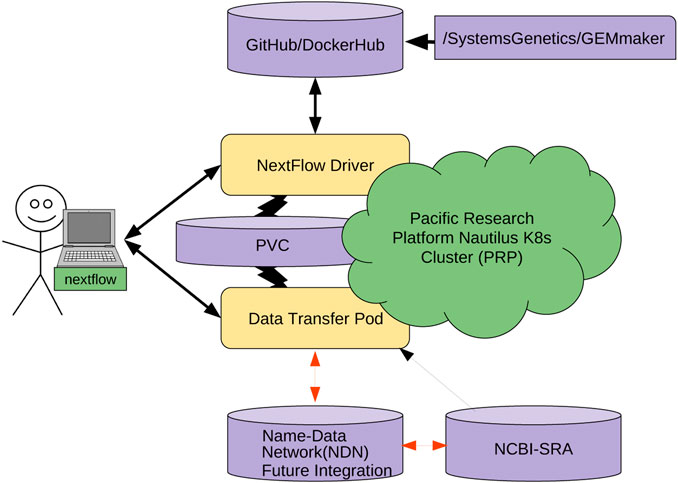
[[ | Advanced [[Imaging informatics|imaging]] and [[DNA sequencing]] technologies now enable the diverse biology community to routinely generate and analyze terabytes of high-resolution biological data. The community is rapidly heading toward the petascale in single-investigator [[laboratory]] settings. As evidence, the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) central DNA sequence repository alone contains over 45 petabytes of biological data. Given the geometric growth of this and other [[genomics]] repositories, an exabyte of mineable biological data is imminent. The challenges of effectively utilizing these datasets are enormous, as they are not only large in size but also stored in various geographically distributed repositories such as those hosted by the NCBI, as well as in the DNA Data Bank of Japan (DDBJ), European Bioinformatics Institute (EBI), and NASA’s GeneLab. ('''[[Journal:Named data networking for genomics data management and integrated workflows|Full article...]]''')<br /> | ||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
{{flowlist | | {{flowlist | | ||
* [[Journal:Implement an international interoperable PHR by FHIR: A Taiwan innovative application|Implement an international interoperable PHR by FHIR: A Taiwan innovative application]] | |||
* [[Journal:Towards a risk catalog for data management plans|Towards a risk catalog for data management plans]] | * [[Journal:Towards a risk catalog for data management plans|Towards a risk catalog for data management plans]] | ||
* [[Journal:Kadi4Mat: A research data infrastructure for materials science|Kadi4Mat: A research data infrastructure for materials science]] | * [[Journal:Kadi4Mat: A research data infrastructure for materials science|Kadi4Mat: A research data infrastructure for materials science]] | ||
}} | }} | ||
Revision as of 17:14, 10 January 2022
"Named data networking for genomics data management and integrated workflows"
Advanced imaging and DNA sequencing technologies now enable the diverse biology community to routinely generate and analyze terabytes of high-resolution biological data. The community is rapidly heading toward the petascale in single-investigator laboratory settings. As evidence, the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) central DNA sequence repository alone contains over 45 petabytes of biological data. Given the geometric growth of this and other genomics repositories, an exabyte of mineable biological data is imminent. The challenges of effectively utilizing these datasets are enormous, as they are not only large in size but also stored in various geographically distributed repositories such as those hosted by the NCBI, as well as in the DNA Data Bank of Japan (DDBJ), European Bioinformatics Institute (EBI), and NASA’s GeneLab. (Full article...)
Recently featured: