Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text) |
Shawndouglas (talk | contribs) (Updated article of the week text) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:GA | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:GA Terlouw JofCheminfo22 14.png|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:PIKAChU: A Python-based informatics kit for analyzing chemical units|PIKAChU: A Python-based informatics kit for analyzing chemical units]]"''' | ||
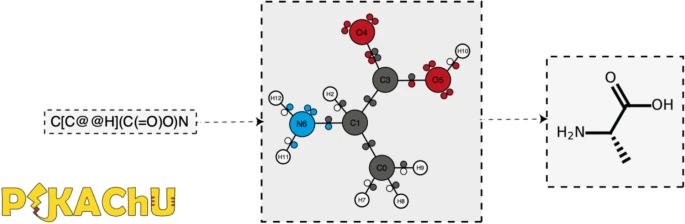
As efforts to computationally describe and simulate the biochemical world become more commonplace, computer programs that are capable of ''in silico'' chemistry play an increasingly important role in biochemical research. While such programs exist, they are often dependency-heavy, difficult to navigate, or not written in [[Python (programming language)|Python]], the programming language of choice for [[Bioinformatics|bioinformaticians]]. Here, we introduce PIKAChU (Python-based Informatics Kit for Analysing CHemical Units), a [[Chemical informatics|cheminformatics]] toolbox with few dependencies implemented in Python. PIKAChU builds comprehensive molecular graphs from simplified molecular-input line-entry system (SMILES) strings, which allow for easy downstream analysis and visualization of molecules. ... ('''[[Journal:PIKAChU: A Python-based informatics kit for analyzing chemical units|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
{{flowlist | | {{flowlist | | ||
* [[Journal:Development of Biosearch System for biobank management and storage of disease-associated genetic information|Development of Biosearch System for biobank management and storage of disease-associated genetic information]] | |||
* [[Journal:Establishing a common nutritional vocabulary: From food production to diet|Establishing a common nutritional vocabulary: From food production to diet]] | * [[Journal:Establishing a common nutritional vocabulary: From food production to diet|Establishing a common nutritional vocabulary: From food production to diet]] | ||
* [[Journal:Designing a knowledge management system for naval materials failures|Designing a knowledge management system for naval materials failures]] | * [[Journal:Designing a knowledge management system for naval materials failures|Designing a knowledge management system for naval materials failures]] | ||
}} | }} | ||
Revision as of 15:55, 20 March 2023
"PIKAChU: A Python-based informatics kit for analyzing chemical units"
As efforts to computationally describe and simulate the biochemical world become more commonplace, computer programs that are capable of in silico chemistry play an increasingly important role in biochemical research. While such programs exist, they are often dependency-heavy, difficult to navigate, or not written in Python, the programming language of choice for bioinformaticians. Here, we introduce PIKAChU (Python-based Informatics Kit for Analysing CHemical Units), a cheminformatics toolbox with few dependencies implemented in Python. PIKAChU builds comprehensive molecular graphs from simplified molecular-input line-entry system (SMILES) strings, which allow for easy downstream analysis and visualization of molecules. ... (Full article...)
Recently featured: