Difference between revisions of "Template:Article of the week"
Shawndouglas (talk | contribs) (Updated article of the week text.) |
Shawndouglas (talk | contribs) (Updated article of the week text.) |
||
| Line 1: | Line 1: | ||
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 | <div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig1 Hong Database2016 2016.jpg|240px]]</div> | ||
'''"[[Journal: | '''"[[Journal:Principles of metadata organization at the ENCODE data coordination center|Principles of metadata organization at the ENCODE data coordination center]]"''' | ||
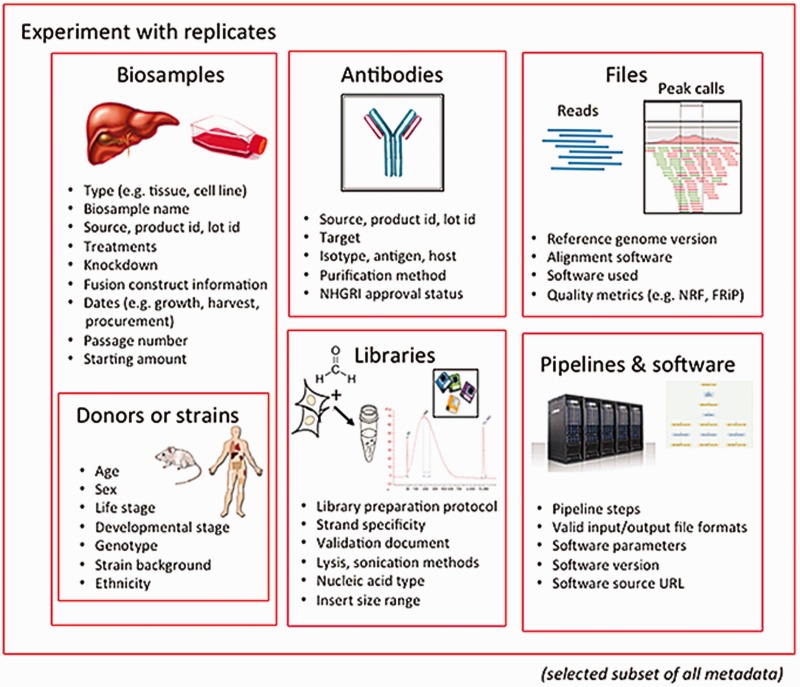
The Encyclopedia of DNA Elements (ENCODE) Data Coordinating Center (DCC) is responsible for organizing, describing and providing access to the diverse data generated by the ENCODE project. The description of these data, known as metadata, includes the biological sample used as input, the protocols and assays performed on these samples, the data files generated from the results and the computational methods used to analyze the data. Here, we outline the principles and philosophy used to define the ENCODE metadata in order to create a metadata standard that can be applied to diverse assays and multiple genomic projects. In addition, we present how the data are validated and used by the ENCODE DCC in creating the ENCODE Portal. ('''[[Journal:Principles of metadata organization at the ENCODE data coordination center|Full article...]]''')<br /> | |||
<br /> | <br /> | ||
''Recently featured'': | ''Recently featured'': | ||
: ▪ [[Journal:Integrated systems for NGS data management and analysis: Open issues and available solutions|Integrated systems for NGS data management and analysis: Open issues and available solutions]] | |||
: ▪ [[Journal:Practical approaches for mining frequent patterns in molecular datasets|Practical approaches for mining frequent patterns in molecular datasets]] | : ▪ [[Journal:Practical approaches for mining frequent patterns in molecular datasets|Practical approaches for mining frequent patterns in molecular datasets]] | ||
: ▪ [[Journal:Improving the creation and reporting of structured findings during digital pathology review|Improving the creation and reporting of structured findings during digital pathology review]] | : ▪ [[Journal:Improving the creation and reporting of structured findings during digital pathology review|Improving the creation and reporting of structured findings during digital pathology review]] | ||
Revision as of 18:19, 7 November 2016
"Principles of metadata organization at the ENCODE data coordination center"
The Encyclopedia of DNA Elements (ENCODE) Data Coordinating Center (DCC) is responsible for organizing, describing and providing access to the diverse data generated by the ENCODE project. The description of these data, known as metadata, includes the biological sample used as input, the protocols and assays performed on these samples, the data files generated from the results and the computational methods used to analyze the data. Here, we outline the principles and philosophy used to define the ENCODE metadata in order to create a metadata standard that can be applied to diverse assays and multiple genomic projects. In addition, we present how the data are validated and used by the ENCODE DCC in creating the ENCODE Portal. (Full article...)
Recently featured: