File:Fig2 Schulz JofPathInformatics2016 7.jpg
Original file (1,210 × 342 pixels, file size: 151 KB, MIME type: image/jpeg)
Summary
| Description |
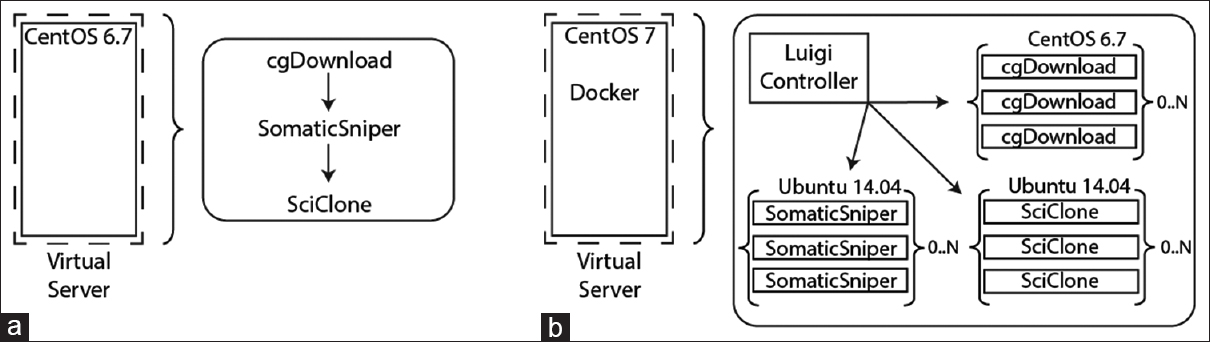
Figure 2. Comparison of standard application architecture and containerized architecture for clonal analysis. (a) When deployed in a virtual server, the analysis workflow was installed on CentOS 6.7 and had to be run serially due to limitations in software parallelization and local resources. Applications are launched manually in sequence to download NGS data, identify variants, and predict tumor clonality. (b) When configured in Docker containers and driven by a workflow manager, applications were automatically launched and able to scale based on available system resources. Each application was configured on its native operating system architecture within the container, as indicated in the figure. |
|---|---|
| Source |
Schulz, W.L.; Durant, T.; siddon, A.J.; Torres, R. (2016). "Use of application containers and workflows for genomic data analysis". Journal of Pathology Informatics 7: 53. doi:10.4103/2153-3539.197197. |
| Date |
2016 |
| Author |
Schulz, W.L.; Durant, T.; siddon, A.J.; Torres, R. |
| Permission (Reusing this file) |
Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported |
| Other versions |
Licensing
|
|
This work is licensed under the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. |
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 00:35, 17 January 2017 | 1,210 × 342 (151 KB) | Shawndouglas (talk | contribs) |
You cannot overwrite this file.
File usage
The following page uses this file:









