File:Fig4 Schulz JofPathInformatics2016 7.jpg
Original file (1,210 × 656 pixels, file size: 123 KB, MIME type: image/jpeg)
Summary
| Description |
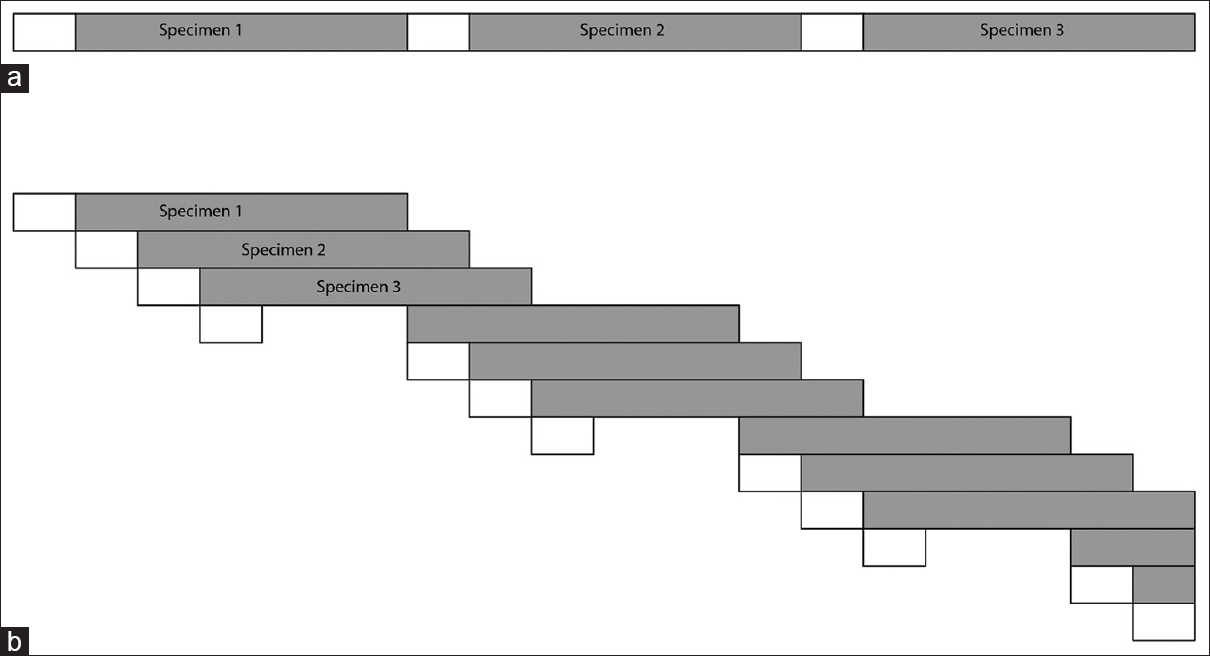
Figure 4. Illustration of parallelization improvements with a workflow-driven container architecture. (a) When performed serially, the download (white bars) and analysis (shaded bars) of a single pair of tumor and germline sequence on local hardware took approximately four hours (bars drawn to scale). (b) When parallelized with a workflow manager and Docker containers, multiple specimens could be processed simultaneously to take advantage of all system resources, including network, memory, and processor capacity. |
|---|---|
| Source |
Schulz, W.L.; Durant, T.; siddon, A.J.; Torres, R. (2016). "Use of application containers and workflows for genomic data analysis". Journal of Pathology Informatics 7: 53. doi:10.4103/2153-3539.197197. |
| Date |
2016 |
| Author |
Schulz, W.L.; Durant, T.; siddon, A.J.; Torres, R. |
| Permission (Reusing this file) |
Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported |
| Other versions |
Licensing
|
|
This work is licensed under the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. |
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 01:04, 17 January 2017 |  | 1,210 × 656 (123 KB) | Shawndouglas (talk | contribs) |
You cannot overwrite this file.
File usage
The following page uses this file:









