Journal:Broad-scale genetic diversity of Cannabis for forensic applications
| Full article title | Broad-scale genetic diversity of Cannabis for forensic applications |
|---|---|
| Journal | PLOS ONE |
| Author(s) | Dufresnes, Christophe; Jan, Catherine; Bienert, Friederike; Goudet, Jérôme; Fumagalli, Luca |
| Author affiliation(s) | University of Lausanne, Centre Universitaire Romand de Médecine Légale, |
| Primary contact | Email: Luca dot Fumagalli at unil dot ch |
| Editors | Scali, Monica |
| Year published | 2017 |
| Volume and issue | 121 |
| Page(s) | e0170522 |
| DOI | 10.1371/journal.pone.0170522 |
| ISSN | 1932-6203 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0170522 |
| Download | http://journals.plos.org/plosone/article/file?id=10.1371/journal.pone.0170522&type=printable (PDF) |
Abstract
Cannabis (hemp and marijuana) is an iconic yet controversial crop. On the one hand, it represents a growing market for pharmaceutical and agricultural sectors. On the other hand, plants synthesizing the psychoactive tetrahydrocannabinol (THC) produce the most widespread illicit drug in the world. Yet, the difficulty to reliably distinguish between Cannabis varieties based on morphological or biochemical criteria impedes the development of promising industrial programs and hinders the fight against narcotrafficking. Genetics offers an appropriate alternative to characterize drug vs. non-drug Cannabis. However, forensic applications require rapid and affordable genotyping of informative and reliable molecular markers for which a broad-scale reference database, representing both intra- and inter-variety variation, is available. Here we provide such a resource for Cannabis, by genotyping 13 microsatellite loci (STRs) in 1,324 samples selected specifically for fiber (24 hemp varieties) and drug (15 marijuana varieties) production. We showed that these loci are sufficient to capture most of the genome-wide diversity patterns recently revealed by next-generation sequencing (NGS) data. We recovered strong genetic structure between marijuana and hemp and demonstrated that anonymous samples can be confidently assigned to either plant types. Fibers appear genetically homogeneous whereas drugs show low (often clonal) diversity within varieties, but very high genetic differentiation between them, likely resulting from breeding practices. Based on an additional test dataset that includes samples from 41 local police seizures, we showed that the genetic signature of marijuana cultivars could be used to trace crime scene evidence. To date, our study provides the most comprehensive genetic resource for Cannabis forensics worldwide.
Introduction
Cannabis is one of humanity’s oldest cultivated plant. It is thought to have originated in central Asia and was domesticated as early as 8,000 BP for food, fiber, oil, medicines and as an inebriant. This crop was since distributed across the world during the last two millennia and, due to its recent legalization in several countries, is increasingly exploited by several industrial sectors (hemp) and as a recreational drug (marijuana). The taxonomic status of Cannabis has always been disputed, as it encompasses multiple cultural, geographic, historical, and functional aspects.[1][2][3][4] Whereas most authors now consider it a monotypic panmictic taxon, Cannabis sativa, three species or subspecies (sativa, indica and ruderalis) are often mentioned but without a comprehensive taxonomic grouping so far. The nomenclature may thus differ depending on whether it refers to morphological or chemical variation, geographic distribution, ecotype, as well as crop-use characteristics and intoxicant properties resulting from human selection.[4][5][6][7] Cannabis presumably diversified following selection for traits enhancing fiber and seed production (”hemp”) or psychoactive properties ("drug"). Importantly, Cannabis types differ in their absolute and relative amounts of terpenophenolic cannabinoids, notably Δ1-tetrahydrocannabinol (THC), the well-known psychoactive compound of marijuana, and the non-psychoactive cannabidiol (CBD). In this context, drug-type Cannabis (marijuana) is broadly characterized by a higher overall cannabinoid content than fiber-types. However, the most widely recognized criteria to assign a Cannabis plant to either “drug” or “hemp” type is the THC:CBD ratio, according to which three main chemical phenotype (chemotype) classes are recognized: hemp-type plants with a low ratio (THC:CBD < 1), drug-type plants with a high ratio (THC:CBD > 1), and intermediate-type plants with a ratio close to one.[6][8] The informal designation sativa and indica may have various, controversial meanings. Morphologically, the name sativa designates tall plants with narrow leaves, while indica refers to short plants with wide leaves. Among the marijuana community however, sativa rather refers to equatorial varieties producing stimulating psychoactive effects (THC:CBD ≈ 1), whereas indica-type plants from Central Asia are used for relaxing and sedative drugs (THC:CBD > 1).[8]
The commercial interest for Cannabis declined during the twentieth century due, e.g., to the development of synthetic fibers and the stringent policies regarding its exploitation, but this iconic weed is recently regaining attention in many countries for its high medicinal, industrial, and agricultural potentials.[9] However, its usage is still controversial, in particular from agro-economic, public health, and forensic perspectives. Due to its intoxicant properties, the cultivation and possession of Cannabis is under strict legal regulations. High-THC:CBD varieties are prohibited in many countries but remain the most frequently-used illicit drug worldwide[10] (~180 million consumers in 2013[11]), in the form of marijuana (dried inflorescences) or hashish (resin). In contrast, low-THC:CBD hemp crops can be exploited under licensed control for seed oil, fibers, and pharmaceuticals. For instance, quantitative measures of THC content are currently considered by the European Union (EU) for approval as a licensed hemp cultivar (below 0.2% THC weight per weight in the mature dry inflorescences; http://ec.europa.eu/food/plant_en). Yet hemp and marijuana varieties are hardly distinguishable morphologically, and discrimination of drug vs. non-drug chemotypes by quantitative THC dosage has also proven inadequate due to its dependence on environmental factors, to the strong variation during the plant’s life cycle, as well as between individual plants.[12][13] In addition, the qualitative assessment of THC:CBD ratio is also problematic for an unequivocal discrimination between fiber and drug types due to the presence of a largely variable intermediate chemotype class, the occurrence of several exceptions (e.g., hemp accessions with a THC-predominant chemotype[14][15][16]), and the common practice among drug breeders to produce hybrid varieties.
This issue largely impedes crops’ improvement and full-scale industrial development; it even causes a security risk, as licensed crops may be used as a cover for illegal drug production. Moreover, it significantly limits the ability of law enforcement agencies to trace drug seizures and link illegal producers to organized crime syndicates supplying the black market of Cannabis drugs. In addition, Cannabis can have long-distance dispersal capabilities[17], and fiber crops might face cryptic contamination by pollen from drug varieties.
Genetic tools offer a promising avenue to overcome these issues, especially to distinguish between drug vs. non-drug plants.[18] Importantly, genetics requires small amounts of tissues as a DNA source, whereas chemical analyses necessitate inflorescences. A promising aspect has been to genotype loci directly linked to THC synthesis[8][19] in association with chemotype profiling. However, this association is not ubiquitous[14][15], and genotyping may be compromised by complex gene duplications, pseudogenes[20][21][22], and the fact that only a limited number of varieties among the tremendous Cannabis diversity has been validated[15]; moreover, chemotype seem to greatly vary even among genotypes.[20]
A parallel, complementary approach is to discriminate drug vs. hemp plants from their non-adaptive genetic variation. Until the recent past, the genetic diversity of Cannabis has remained surprisingly under-investigated, partly due to the important restrictions imposed by anti-drug policies, even for scientific inquiries. In the last few years, a draft genome of Cannabis was published[22], and high-density Single-Nucleotide-Polymorphism (SNP) data obtained from NGS techniques evidenced genome-wide differentiation between hemp and marijuana plants.[23] However, genetic resources applicable for forensics remain under-developed. Forensic investigations require sets of sufficiently informative loci that can be genotyped in large batches of samples in a rapid and affordable manner, such as microsatellites (Short-Tandem-Repeats, STRs). Another prerequisite is that the species’ diversity is exhaustively represented in reference databases, both within and among varieties, so that investigated samples of unknown origin can be identified with statistical confidence. In Cannabis, these two aspects are challenging given the diversity of varieties, their complex breeding histories, as well as the rapid shifts of the drug varieties available on black markets. In addition, hemp and marijuana diverged during the human era and still largely share a common pool of genetic variation.[23]
Several microsatellite analyses were previously performed on Cannabis. Some loci became available in the early 2000s[24][25][26] but remained scarcely tested at the individual or population level. The first STR multiplex kit for forensics was validated years later[27], and subsequently trialed to distinguish fibers from confiscated drug seizures in Australia, with moderate success.[28] Another STR kit was developed by Köhnemann et al.[29], although without reference data. Using transcriptomic sequences (EST), Gao et al.[30] isolated >100 STRs, allowing them to discriminate between Chinese and European hemp samples according to their geographic origin. Other studies genotyped Cannabis, notably from police seizures, using new or published markers.[31][32][33][34][35] However, although these studies are regionally and timely relevant, they rely on limited sample sets (i.e., few varieties and few individuals per variety, and/or only representing plants available on a regional black market at the time of confiscations), thus hardly accounting for the different levels of genetic variation of Cannabis stocks. So far no comprehensive database of Cannabis diversity exists for broad-scale forensic enquiries.
Considering these limitations, we developed a new STR resource for Cannabis forensics. We analyzed intra- and inter-populational variation at 13 published STR markers in >1,300 Cannabis samples from 48 fiber and drug accessions, broadly representative of known hemp and marijuana varieties (see Table S1 in "Supporting information"), and characterized unknown samples of various origins, notably police seizures. We aimed at (i) showing that these loci fully recover the genetic structure between marijuana and hemp; (ii) demonstrating that anonymous samples can be confidently assigned to either plant types; and (iii) documenting the genetic diversity among and within samples and its potential for forensic investigations.
Results and discussion
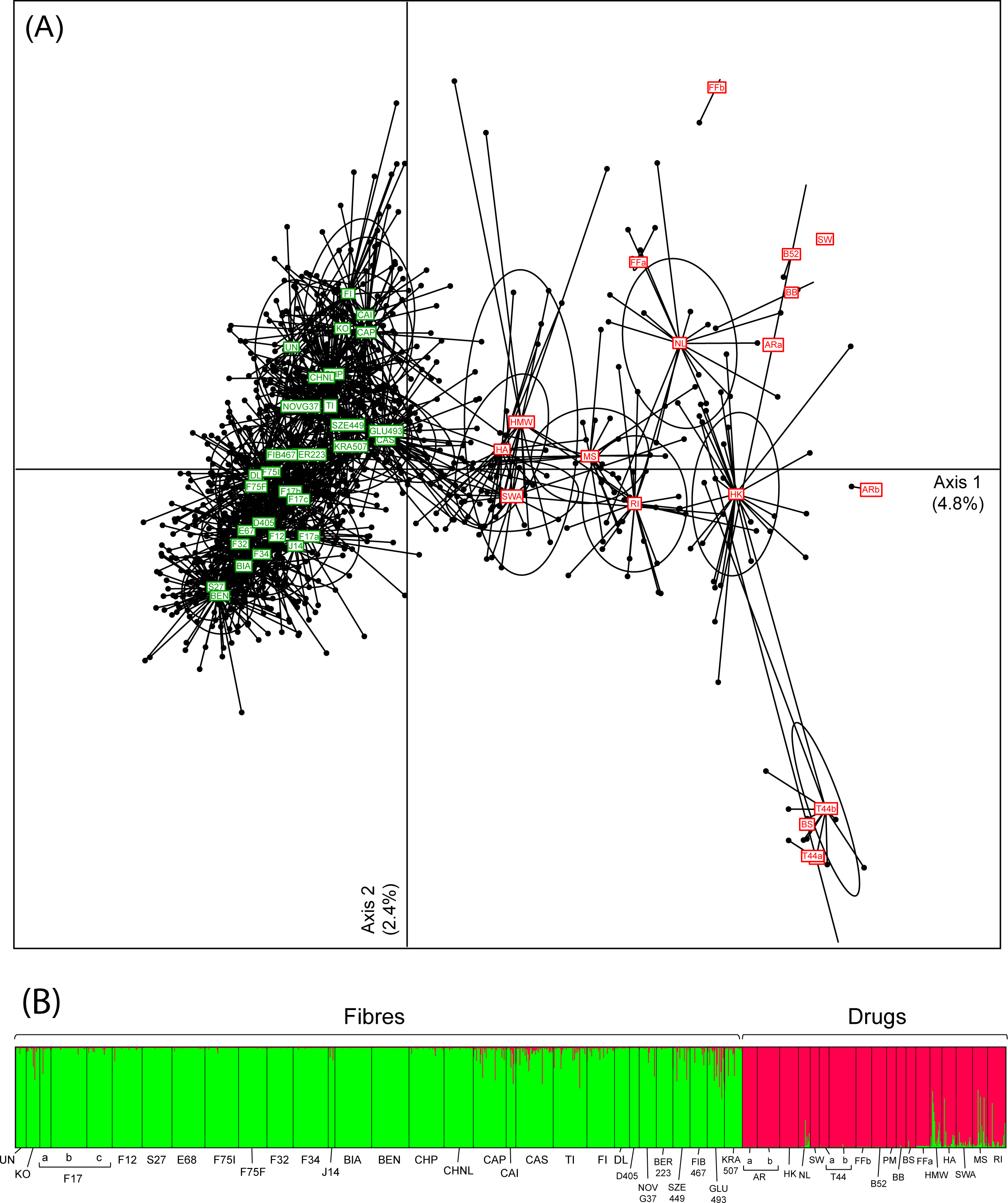
The selected STR markers (see Table S2 in "Supporting information") unanimously recovered the strong structure between fibers and drug Cannabis samples. This is clearly depicted by a principal component analysis (PCA, Fig. 1A), genetic distances between accessions (Fst, Fig. S1 in "Supporting information") and genotype clustering by STRUCTURE (Fig. 1B), where two groups appears as the best clustering solution (ΔK2 = 1205.6). As recently evidenced from NGS data[23], this pattern reflects differentiation between hemp and marijuana over the entire genome, not only at genes underlying THC and fiber synthesis. Some drugs and fibers show weak signs of genetic admixture (intermediate PCA scores and STRUCTURE probabilities, Fig 1; lower Fst, Fig. S1 in "Supporting information"), which might stem from introgressive crossbreeding, as reported elsewhere.[23] Interestingly, except for RI (indica/ruderalis hybrid), all drug varieties closely related to hemps are of sativa ancestry (HMW, HA, SWA, MS; based on available information from suppliers). This would support the common assumption that hemp varieties selected for fiber and seed production derived from sativa, although this view has been challenged by other studies that found more similarities between hemp and indica.[7][23][36] Alternatively, sativa drugs, which are nowadays distributed in more equatorial regions, may be frequently crossbred with indica and agricultural varieties to facilitate their cultivation in temperate countries. In any case, marijuana genetic diversity seems weakly associated with the documented breeding history. We also performed a PCA solely on drugs, which only marginally clustered according to their main sativa and indica pedigree (Fig. S2 in "Supporting information"). Some cultivars of the same appellation appear genetically distinct (e.g., Alpine Rocket, ARa and ARb, FST = 0.36) whereas others harboring different names are genetically identical (e.g., PM, T44, BS, FST = 0.00; identical clones shared by ARa and B52, Table S1 in "Supporting information"). Overall, these observations are in line with the general conclusions of Sawler et al.[23] that drug varieties are often misinformed due to the clandestine nature of Cannabis breeding over the last century, and that names do not necessarily reflect a meaningful genetic identity. In addition, hemp varieties were grouped according to reproductive characteristics, as expected (dioecious versus monoecious; Table S1 in "Supporting information"), as a result of their breeding history (illustrated on the PCA, Fig 1; Fst tree, Fig. S1 in "Supporting information").
|
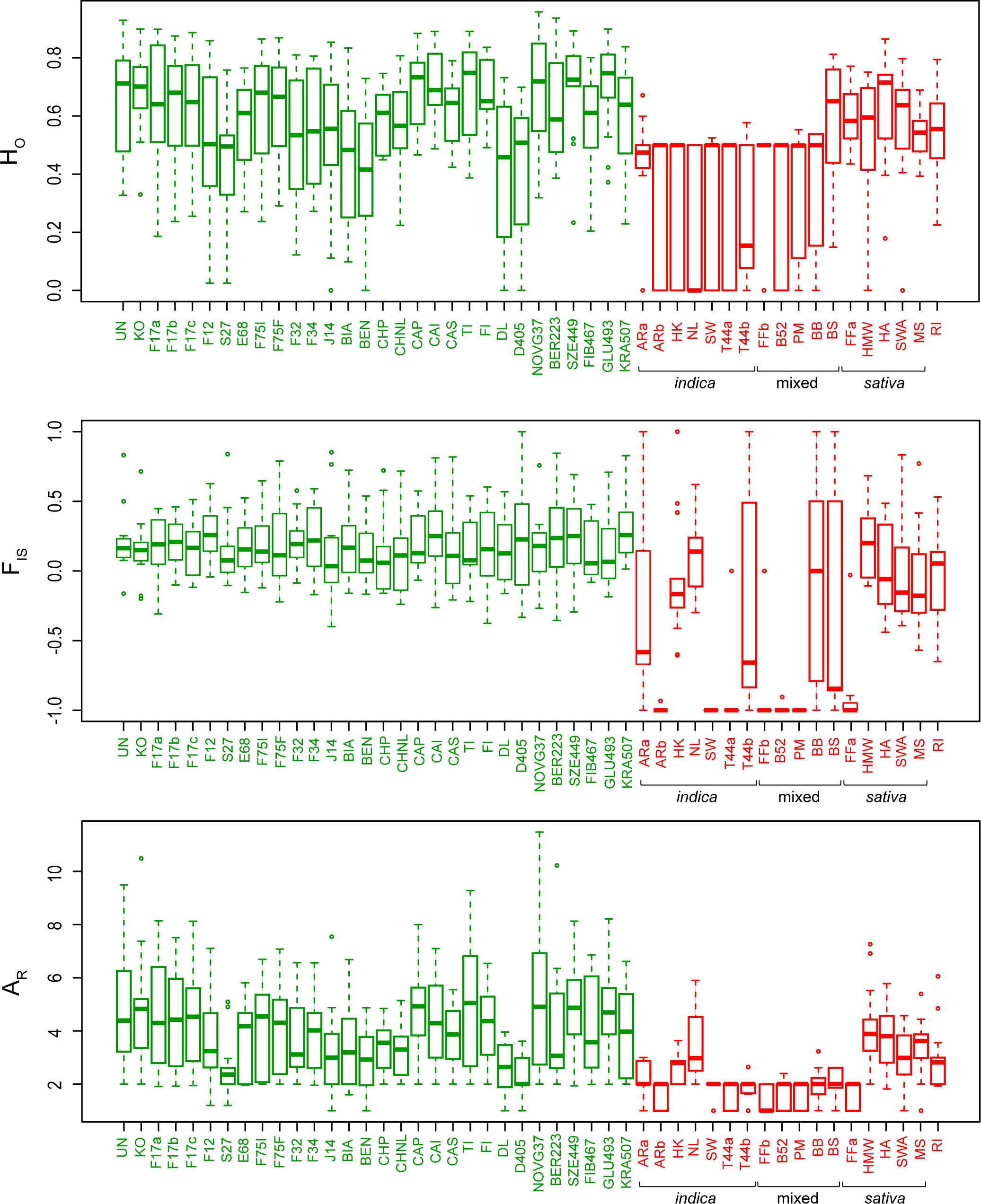
Intra-variety diversity was relatively similar among hemps (Fig. 2). Allelic richness (average number of alleles per population AR, scaled to eight individuals) and heterozygosity (HO) averaged 4.0 ± 0.8 and 0.59 ± 0.10 respectively (Fig. 2). All varieties had positive inbreeding coefficients (FIS = 0.19 ± 0.05), potentially reflecting bottlenecks linked to current breeding practices. The overall differentiation among hemps was relatively low (FST = 0.15 ± 0.07; Fig. S1 in "Supporting information"). In contrast, marijuana featured lower diversity within varieties (AR = 2.3 ± 0.9, HO = 0.41 ± 0.15; Fig. 2) but substantially higher genetic distances among them (FST = 0.39 ± 0.16; Fig. S1 in "Supporting information"). We detected identical genotypes (clones) and strong excess of heterozygosity among several breeds (all of indica or mixed origin, Table S1 in "Supporting information"), which translates into AR of 2, HO of 0.5, and FIS reaching -1 (Fig. 2), resulting from clonal breeding from hybrids of two different parental strains. Interestingly, sativa drugs featured more hemp-like patterns of diversity. Overall, the homogeneous gene pool of hemps suggests more frequent crossbreeding compared to drugs[23], especially of indica content, and/or that a wider genetic base has been sourced by the hemp industry. Marijuana is often propagated clonally for practical reasons as well as to protect the genetic identity of varieties from contamination by wind-dispersing pollens, thus reducing diversity and triggering strong heterozygosity in F1 cross-breeds. Moreover, all Cannabis drug forms are dioecious, and males, which produce lower amounts of THC than females, are discarded by breeders, which further reduces diversity.
|
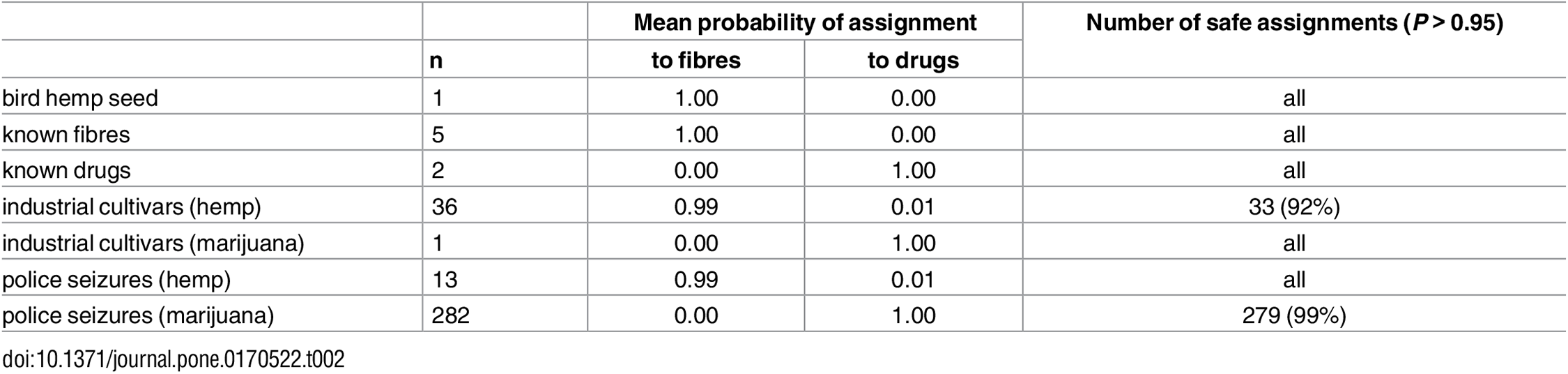
The diversity captured by our STR markers appears well representative of the genomic background of Cannabis: our results are overall very concordant with high-density SNP data.[23] Our STR database thus seems appropriate for broad-scale forensic applications, in particular to discriminate between drug vs. non-drug samples, one of the main tasks of Cannabis forensics. To demonstrate this ability, we performed genetic assignment tests (direct or resampling-based) on random subsets of drug and fiber samples, using the remainder of the dataset as reference (detailed in the Methods section). The direct test always correctly assigned every sample to their plant type (Table 1). The more conservative resampling approach never misassigned any specimen (Table 1). Many individuals are yet not assigned to any group (even the correct one) because genotypes are considered not statistically informative enough by this conservative analysis. We further evaluated the database by genotyping 340 additional Cannabis samples of various origins (bird food, drug and fiber specimens, uncertain industrial cultivars and police seizures). Known specimens (n = 8) were all correctly assigned with high confidence (Table 2). All but one industrial cultivars (n = 37) consisted of hemps, with few getting assignment probabilities below 0.95 (Table 2). Confiscated samples (n = 295, from 41 different seizures) could be unambiguously assigned except for three specimens (Table 2).
|
|
These results clearly illustrate the relevance of our new database for forensics. Notably, it outperforms the reference published by Howard et al.[28] for Australian seizures, which suffered substantial mis-assignment risks, but yet so far was the only available resource properly tested by statistical assignments. Moreover, compared to previous studies, our sampling scheme has the advantage of covering a broad range of Cannabis varieties and accounts for their intra-variety variation. The latter seems important to consider, as some marijuana (sativa) and hemp cultivars share closely related gene pools, sometimes making their discrimination difficult.
In addition, the strong genetic structure between drug cultivars may provide opportunities for police investigations of narcotrafficking. One challenge for law-enforcement agencies is to trace evidence collected at crime scenes in order to connect and convict acting members of crime syndicates. Most marijuana individuals/germlines show unique genetic profiles at our markers (Fig. 1A, Fig S2 in "Supporting information"), so they could be suitable for this task. We screened for identical genotypes among the seized Western-Swiss samples of our test dataset, where the probability of identity PI-sib is 8.9 × 10−5. We could establishe five groups of related seizures (some even matched by several germlines) thus with 99.991% confidence (Table S3 in "Supporting information"); the remaining 25 seizures were genetically different (Table S3 in "Supporting information"). Given the high resolution at such narrow regional scale, this approach could also be applied at national or international levels. The illegal trade of Cannabis is one of the most developed illicit industries in the world (>7,000 tons seized in 2013[11]), yearly generating enormous profits used to finance other criminal activities. Exploiting the genetic heterogeneity of marijuana should be the focus of further forensic development to aid the international fight against narcotrafficking.
To date, our STR database is the most powerful resource suitable for routine forensic analyses of Cannabis. Yet, it remains limited by several aspects. First, drug vs. non-drug discrimination can be ambiguous for some samples, given the lack of differentiation and/or crossbreeding practices between few hemp and marijuana varieties. Second, the plant type of our reference samples rely on the information provided by the suppliers, which could be confirmed by chemotyping analyses. Third, more sensitive applications such as tracing drug evidences might require a finer resolution. In both cases, updating the database with additional markers and reference populations, especially new drug varieties, seems a worthy investment. Further development would benefit from international collaborations. An array of genetic studies have been conducted on Cannabis in just a few years by different research teams (see Introduction), each contributing specific sets of samples and markers. Given the tremendous diversity of marijuana and the legal difficulty to access samples, joint efforts between Cannabis genetics experts worldwide would allow unprecedented opportunities to extend forensic advances and promote the development of the industrial and therapeutic potential of this emblematic species.
Materials and methods
Ethics statement
This study does not involve any endangered or protected species.
Sample collection
We built a collection of 1,324 Cannabis samples from 30 accessions of fibers (n = 972 from 24 different varieties) and 18 accessions of drug (n = 352 from 15 varieties). These accessions broadly cover the legal European hemp varieties (landraces, cultivars selected from landraces, and cross-bred cultivars) and marijuana diversity (identified a priori as sativa, indica, and hybrids by breeders). In order to also capture intra-variety variation, we included large population samples for each accession (27 samples on average, from 9 to 50). Seeds and leaves were obtained from agronomic companies, germplasm collections, police seizures, or commercial stores; seeds were germinated at the University of Lausanne (Switzerland). Table S1 in the "Supporting information" section provides sample origin and reported breeding history, given available documentation and information provided by the suppliers.
To evaluate our reference database, we further considered 340 additional test samples from uncertain (police seizures, industrial cultivars) or known types (fiber and drug samples not included in the reference database). Confiscated plants (n = 295) represented 41 police seizures across Western Switzerland from 2005 to 2010. Details are provided in Table S3 in "Supporting information".
DNA extraction and microsatellite genotyping
DNA was extracted from approximately 25 mg of dried plant leaves using the FastDNA Kit (Qbiogene, Carlsbad, CA) following the manufacturer’s instructions. Thirteen published microsatellite loci were analyzed[24][25], including the 10 from Howard et al.’s forensically validated kit.[28] DNA amplifications were performed according to their STR multiplex system (M1 and M4), slightly modified to include ANUCS202 to multiplex M4. In addition, we integrated a new multiplex M5 to amplify loci ANUCS201 and H09-CANN2. Detailed information on markers and multiplexes are available in Table S2 in "Supporting information.". PCR conditions were as follows: 95°C for 5 minutes (initial denaturation); 10 cycles consisting of 30” at 95°C, 30” at 66°C down to 54°C (-3°C/2 cycles) and 45” at 72°C (top-down PCR); 30 regular PCR cycles consisting of 30” at 95°C, 30” at 50°C and 45” at 72°C; 90”at 72°C (final elongation). Amplicons were run on an ABI PRISM 3130 Genetic Analyzer (Applied Biosystems) and genotyped were scored using GeneMapper v3.2 (ABI).
Population genetic analyses
We analyzed the genetic structure and diversity of Cannabis by three different approaches. First, we performed a principal component analysis (PCA) on individual genotypes using the R packages ade4 and adegenet.[37] Second, we conducted Bayesian clustering of genotypes into groups with STRUCTURE.[38] We used the admixture model without prior on sample origin, testing from one to 11 groups (K), with 10 replicates per K. Each run consisted of 100’000 iterative steps following a burn-in of 10’000. We applied the Evanno method[39] to determine the most likely number of groups summarizing the data, as implemented in STRUCTURE HARVESTER.[40] Replicates were combined using CLUMPP[41] and graphical displays of admixture proportions (barplots) were built with DISTRUCT.[42] Third, we conducted population-based analyses with FSTAT[43], by calculating pairwise genetic distances between accessions (FST) as well as the following diversity indices for each accession: observed heterozygosity (HO), inbreeding coefficient (FIS) and allelic richness (AR, scaled to eight individuals).
Genotype specificity and assignment tests
We used GenAlEx 6[44] to compute, within and among accessions, the number of private alleles (PA) and probabilities of identity (PI), i.e., the probability to have identical genotypes by chance. For the latter, we considered the conservative estimate of PI-sib when the data potentially includes siblings, as appropriate for Cannabis samples. We also used GenAlEx to match identical genotypes, notably to identify clones (function “match”).
To assess the power of discrimination between hemp and drug types, assignment analyses were performed with GeneClass2.[45] First, we auto-evaluated our database by assigning 10 re-sampled random subsets (representing about 10% of the total dataset, n = 100 for fibers, n = 40 for drugs) using the rest of the data as reference. To this end, two different methods proposed by the software were applied, using Bayesian criteria.[46] The first approach (direct method) estimates the proportion of correctly assigned samples to the most likely population of origin. The second approach (resampling-based method) computes the probability that samples belong to each reference population and aims at minimizing the risk of mis-assignment, i.e., when individuals feature genotypes that can occur in the “wrong” reference population (type I error). This was achieved by simulating the likelihood distribution of 10,000 independent genotypes, for each reference population (with a Monte-Carlo resampling algorithm[47]), against which the genotypes to assign can then be compared. Rejection or inclusion is then decided upon a threshold (P < 0.01). This approach does not assume that all source populations have been sampled. Second, we assigned (direct method), our 340 test samples, which consist mostly of unknown varieties.
Supporting information
Fig. S1: Tree of genetic distances (pairwise Fst) between Cannabis accessions (Monoecious hemp are highlighted in grey)
https://doi.org/10.1371/journal.pone.0170522.s001 (TIF)
Fig. S2: Genetic structure among marijuana samples
https://doi.org/10.1371/journal.pone.0170522.s002 (TIF)
Table S1: List and details on the Cannabis accessions
https://doi.org/10.1371/journal.pone.0170522.s003 (XLSX)
Table S2: List and details on the STRs markers
https://doi.org/10.1371/journal.pone.0170522.s004 (XLSX)
Table S3: List and details on test samples
https://doi.org/10.1371/journal.pone.0170522.s005 (XLSX)
Acknowledgements
We thank N. Boschung, P. Busso, N. Duvoisin, J. El Assad, A. Gaigher, L. Gigord, N. Giroud, J. Goebel, K. Ridout, N. Ruech, C. Stoffel for help in the greenhouse and in the laboratory; V. Castella and C. Giroud at Centre Universitaire Romand de Médecine Légale (Lausanne, Switzerland); P. Cantin and the Institut de Police Scientifique (Lausanne, Switzerland), J. Elzinga and A. Hazelkamp for sampling, as well as the Swiss police departments from the cantons of Fribourg, Neuchâtel (in particular O. Guéniat) and Vaud for seizures; S. Grigoryev from the Vavilov Institute of Plant Genetic Resources (St. Petersburg, Russia), Prof. G. Venturi (Bologna University, Italy), and the agronomic companies listed in Table S1 for hemp seeds; the shops listed in Table S1 for marijuana seeds and leaf samples; C. Howard and R. Peakall (Australian National University, Canberra, Australia) for reference samples; D. Jeffries for proof-reading the manuscript.
Author contributions
Conceptualization: JG LF
Formal analysis: CD FB JG LF
Funding acquisition: LF
Investigation: CJ LF
Methodology: CJ LF
Project administration: LF
Supervision: LF
Validation: CJ LF
Visualization: CD LF
Writing – original draft: CD LF
Writing – review & editing: CD CJ FB JG LF
Additional notes
Data availability
Microsatellite genotypes are available from the Dryad Digital Repository (doi:10.5061/dryad.p2d8h).
Funding
This work was supported by Swiss National Science Foundation (SNSF). Grant number 31003A_130234 to LF. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing interests
The authors have declared that no competing interests exist.
References
- ↑ Small, E.; Crongquist, A. (1976). "A practical and natural taxonomy for cannabis". Taxon 25 (4): 405–435. doi:10.2307/1220524.
- ↑ Clarke, R.C.; Merlin, M.D. (2013). Cannabis: Evolution and Ethnobotany. University of California Press. pp. 434. ISBN 9780520270480.
- ↑ Small, E. (2015). "Evolution and Classification of Cannabis sativa (Marijuana, Hemp) in Relation to Human Utilization". The Botanical Review 81 (3): 189–294. doi:10.1007/s12229-015-9157-3.
- ↑ 4.0 4.1 Welling, M.T.; Shapter, T.; Rose, T.J. et al. (2016). "A Belated Green Revolution for Cannabis: Virtual Genetic Resources to Fast-Track Cultivar Development". Frontiers in Plant Science 7: 1113. doi:10.3389/fpls.2016.01113. PMC PMC4965456. PMID 27524992. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4965456.
- ↑ de Meijer, E.P.M.; van Soest, L.J.M. (1992). "The CPRO Cannabis germplasm collection". Euphytica 62 (3): 201–11. doi:10.1007/BF00041754.
- ↑ 6.0 6.1 de Meijer, E.P.M. (2014). "The Chemical Phenotypes (Chemotypes) of Cannabis". In Pertwee, R.. Handbook of Cannabis. Oxford University Press. pp. 89–110. ISBN 9780199662685.
- ↑ 7.0 7.1 Hillig, K.W. (2005). "Genetic evidence for speciation in Cannabis (Cannabaceae)". Genetic Resources and Crop Evolution 52 (2): 161–80. doi:10.1007/s10722-003-4452-y.
- ↑ 8.0 8.1 8.2 Hillig, K.W.; Mahlberg, P.G. (2004). "A chemotaxonomic analysis of cannabinoid variation in Cannabis (Cannabaceae)". American Journal of Botany 91 (6): 966–75. doi:10.3732/ajb.91.6.966. PMID 21653452.
- ↑ Andre, C.M.; Hausman, J.F.; Guerriero, G. (2016). "Cannabis sativa: The Plant of the Thousand and One Molecules". Frontiers in Plant Science 7: 19. doi:10.3389/fpls.2016.00019. PMC PMC4740396. PMID 26870049. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4740396.
- ↑ Anderson, P. (2006). "Global use of alcohol, drugs and tobacco". Drug and alcohol review 25 (6): 489–502. doi:10.1080/09595230600944446. PMID 17132569.
- ↑ 11.0 11.1 United Nations Office on Drugs and Crime (2015) (PDF). World Drug Report 2015. United Nations. pp. 162. ISBN 9789211482829. https://www.unodc.org/documents/wdr2015/World_Drug_Report_2015.pdf.
- ↑ Rowan, M.G.; Fairbairn, J.W. (1977). "Cannabinoid patterns in seedlings of Cannabis sativa L. and their use in the determination of chemical race". Journal of Pharmacy and Pharmacology 29 (8): 491–4. PMID 19599.
- ↑ Baker, P.B.; Gough, T.A.; Taylor, B.J. (1982). "The physical and chemical features of Cannabis plants grown in the United Kingdom of Great Britain and Northern Ireland from seeds of known origin". Bulletin of Narcotics 34 (1): 27-36. PMID 6291677.
- ↑ 14.0 14.1 Welling, M.T.; Liu, L.; Shapter, T. et al. (2016). "Characterisation of cannabinoid composition in a diverse Cannabis sativa L. germplasm collection". Euphytica 208 (3): 463–75. doi:10.1007/s10681-015-1585-y.
- ↑ 15.0 15.1 15.2 Staginnus, C.; Zörntlein, S.; de Meijer, E. (2014). "A PCR marker linked to a THCA synthase polymorphism is a reliable tool to discriminate potentially THC-rich plants of Cannabis sativa L.". Journal of Forensic Sciences 59 (4): 919-26. doi:10.1111/1556-4029.12448. PMID 24579739.
- ↑ Tipparat, P.; Natakankitkul, S.; Chamnivikaipong, P.; Chutiwat, S. (2012). "Characteristics of cannabinoids composition of Cannabis plants grown in Northern Thailand and its forensic application". Forensic Science International 215 (1–3): 164-70. doi:10.1016/j.forsciint.2011.05.006. PMID 21636228.
- ↑ Cabezudo, B.; Recio, M.; Sánchez-Laulhé, J. et al. (1997). "Atmospheric transportation of marihuana pollen from North Africa to the Southwest of Europe". Atmospheric Environment 31 (20): 3323-3328. doi:10.1016/S1352-2310(97)00161-1.
- ↑ Miller Coyle, H.; Palmbach, T.; Juliano, N. et al. (2003). "An overview of DNA methods for the identification and individualization of marijuana". Croatian Medical Journal 44 (3): 315–21. PMID 12808725.
- ↑ de Meijer, E.P.; Bagatta, M.; Carboni, A. et al. (2003). "The inheritance of chemical phenotype in Cannabis sativa L.". Genetics 163 (1): 335–46. PMC PMC1462421. PMID 12586720. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1462421.
- ↑ 20.0 20.1 Weiblen, G.D.; Wenger, J.P.; Craft, K.J. et al. (2015). "Gene duplication and divergence affecting drug content in Cannabis sativa". The New Phytologist 208 (4): 1241–50. doi:10.1111/nph.13562. PMID 26189495.
- ↑ McKernan, K.J.; Helbert, Y.; Tadigotla, V. et al. (2015). "Single molecule sequencing of THCA synthase reveals copy number variation in modern drug-type Cannabis sativa L.". bioRxiv. doi:10.1101/028654.
- ↑ 22.0 22.1 van Bakel, H.; Stout, J.M.; Cote, A.G. et al. (2011). "The draft genome and transcriptome of Cannabis sativa". Genome Biology 12 (10): R102. doi:10.1186/gb-2011-12-10-r102. PMC PMC3359589. PMID 22014239. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3359589.
- ↑ 23.0 23.1 23.2 23.3 23.4 23.5 23.6 23.7 Sawler, J.; Stout, J.M.; Gardner, K.M. et al. (2015). "The Genetic Structure of Marijuana and Hemp". PLOS ONE 10 (8): e0133292. doi:10.1371/journal.pone.0133292. PMC PMC4550350. PMID 26308334. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4550350.
- ↑ 24.0 24.1 Alghanim, H.J.; Almirall, J.R. (2003). "Development of microsatellite markers in Cannabis sativa for DNA typing and genetic relatedness analyses". Analytical and Bioanalytical Chemistry 376 (8): 1225-33. doi:10.1007/s00216-003-1984-0. PMID 12811461.
- ↑ 25.0 25.1 Gilmore, S.; Peakall, R.; Robertson, J. (2003). "Short tandem repeat (STR) DNA markers are hypervariable and informative in Cannabis sativa: Implications for forensic investigations". Forensic Science International 131 (1): 65-74. PMID 12505473.
- ↑ Hsieh, H.M.; Hou, R.J.; Tsai, L.C. et al. (2003). "SA highly polymorphic STR locus in Cannabis sativa". Forensic Science International 131 (1): 53–8. PMID 12505471.
- ↑ Howard, C.; Gilmore, S.; Robertson, J. et al. (2008). "Developmental validation of a Cannabis sativa STR multiplex system for forensic analysis". Journal of Forensic Sciences 53 (5): 1061-7. doi:10.1111/j.1556-4029.2008.00792.x. PMID 18624889.
- ↑ 28.0 28.1 28.2 Howard, C.; Gilmore, S.; Robertson, J. et al. (2009). "A Cannabis sativa STR genotype database for Australian seizures: Forensic applications and limitations". Journal of Forensic Sciences 54 (3): 556-63. doi:10.1111/j.1556-4029.2009.01014.x. PMID 19302382.
- ↑ Köhnemann, S.; Nedele, J.; Schwotzer, D. et al. (2012). "The validation of a 15 STR multiplex PCR for Cannabis species". International Journal of Legal Medicine 126 (4): 601–6. doi:10.1007/s00414-012-0706-6. PMID 22573357.
- ↑ Gao, C.; Xin, P.; Cheng, C. et al. (2014). "Diversity analysis in Cannabis sativa based on large-scale development of expressed sequence tag-derived simple sequence repeat markers". PLOS ONE 9 (10): e110638. doi:10.1371/journal.pone.0110638. PMC PMC4203809. PMID 25329551. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4203809.
- ↑ Chandra, S.; Lata, H.; Techen, N. et al. (2011). "Analysis of Genetic Diversity using SSR Markers and Cannabinoid Contents in Different Varieties of Cannabis sativa L.". Planta Medica 77: P_5. doi:10.1055/s-0031-1273534.
- ↑ Valverde, L.; Lischka, C.; Scheiper, S. et al. (2014). "Characterization of 15 STR cannabis loci: Nomenclature proposal and SNPSTR haplotypes". Forensic Science International Genetics 9: 61–5. doi:10.1016/j.fsigen.2013.11.001. PMID 24528581.
- ↑ Valverde, L.; Lischka, C.; Erlemann, S. et al. (2014). "Nomenclature proposal and SNPSTR haplotypes for 7 new Cannabis sativa L. STR loci". Forensic Science International Genetics 13: 185–6. doi:10.1016/j.fsigen.2014.08.002. PMID 25173491.
- ↑ Presinszka, M.; Stiasna, K.; Vyhnanek, T. et al. (2015). "Analysis of microsatellite markers in hemp (Cannabis sativa L.)". In Polák, O.; Cerkal, R.; Belcredi, N.B. (PDF). MendelNet 2015: Proceedings of International PhD Students Conference. Mendel University in Brno. pp. 434–438. ISBN 9788075093639. https://mnet.mendelu.cz/mendelnet2015/mnet_2015_full.pdf.
- ↑ Houston, R.; Birck, M.; Hughes–Stamm, S. et al. (2016). "Evaluation of a 13-loci STR multiplex system for Cannabis sativa genetic identification". International Journal of Legal Medicine 130 (3): 635-47. doi:10.1007/s00414-015-1296-x. PMID 26661945.
- ↑ Pilluza, G.; Delogu, G.; Cabras, A. et al. (2013). "Differentiation between fiber and drug types of hemp (Cannabis sativa L.) from a collection of wild and domesticated accessions". Genetic Resources and Crop Evolution 60 (8): 2331–2342. doi:10.1007/s10722-013-0001-5.
- ↑ Jombart, T. (2008). "adegenet: A R package for the multivariate analysis of genetic markers". Bioinformatics 24 (11): 1403–5. doi:10.1093/bioinformatics/btn129. PMID 18397895.
- ↑ Pritchard, J.K.; Stephens, M.; Donnelly, P. (2000). "Inference of population structure using multilocus genotype data". Genetics 155 (2): 945-59. PMC PMC1461096. PMID 10835412. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1461096.
- ↑ Evanno, G.; Regnaut, S.; Goudet, J. (2005). "Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study". Molecular Ecology 14 (8): 2611–20. doi:10.1111/j.1365-294X.2005.02553.x. PMID 15969739.
- ↑ Earl, D.A.; von Holdt, B.M. (2012). "STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method". Conservation Genetics Resources 4 (2): 359–361. doi:10.1007/s12686-011-9548-7.
- ↑ Jakobsson, M.; Rosenberg, N.A. (2007). "CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure". Bioinformatics 23 (14): 1801-6. doi:10.1093/bioinformatics/btm233. PMID 17485429.
- ↑ Rosenberg, N.A. (2004). "distruct: A program for the graphical display of population structure". Molecular Ecology Resources 4 (1): 137-138. doi:10.1046/j.1471-8286.2003.00566.x.
- ↑ Goudet, J. (1995). "FSTAT (Version 1.2): A Computer Program to Calculate F-Statistics". Journal of Heredity 86 (6): 485–486. doi:10.1093/oxfordjournals.jhered.a111627.
- ↑ Peakall, R.; Smouse, P.E. (2012). "GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research--an update". Bioinformatics 28 (19): 2537-9. doi:10.1093/bioinformatics/bts460. PMC PMC3463245. PMID 22820204. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3463245.
- ↑ Piry, S.; Alapetite, A.; Cornnuet, J.M. et al. (2004). "GENECLASS2: A software for genetic assignment and first-generation migrant detection". Journal of Heredity 95 (6): 536-9. doi:10.1093/jhered/esh074. PMID 15475402.
- ↑ Rannala, B.; Mountain, J.L. (1997). "Detecting immigration by using multilocus genotypes". Proceedings of the National Academy of Sciences of the United States of America 94 (17): 9197-201. PMC PMC23111. PMID 9256459. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC23111.
- ↑ Paetkau, D.; Slade, R.; Burden, M. (2004). "Genetic assignment methods for the direct, real-time estimation of migration rate: A simulation-based exploration of accuracy and power". Molecular Ecology 13 (1): 55–65. PMID 14653788.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added.