File:Fig1 Schulz JofPathInformatics2016 7.jpg
Fig1_Schulz_JofPathInformatics2016_7.jpg (705 × 473 pixels, file size: 89 KB, MIME type: image/jpeg)
Summary
| Description |
Figure 1. Serial workflow and architecture to download Cancer Genomics Hub data. To obtain next generation sequencing data from Cancer Genomics Hub, the cgDownload utility was used to transfer aligned whole genome and whole exome sequencing data. The SomaticSniper utility was then used to identify somatic variants and tumor clonality was predicted with SciClone. These utilities were all manually configured on a server running CentOS 6.7 |
|---|---|
| Source |
Schulz, W.L.; Durant, T.; siddon, A.J.; Torres, R. (2016). "Use of application containers and workflows for genomic data analysis". Journal of Pathology Informatics 7: 53. doi:10.4103/2153-3539.197197. |
| Date |
2016 |
| Author |
Schulz, W.L.; Durant, T.; siddon, A.J.; Torres, R. |
| Permission (Reusing this file) |
Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported |
| Other versions |
Licensing
|
|
This work is licensed under the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. |
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 00:22, 17 January 2017 | 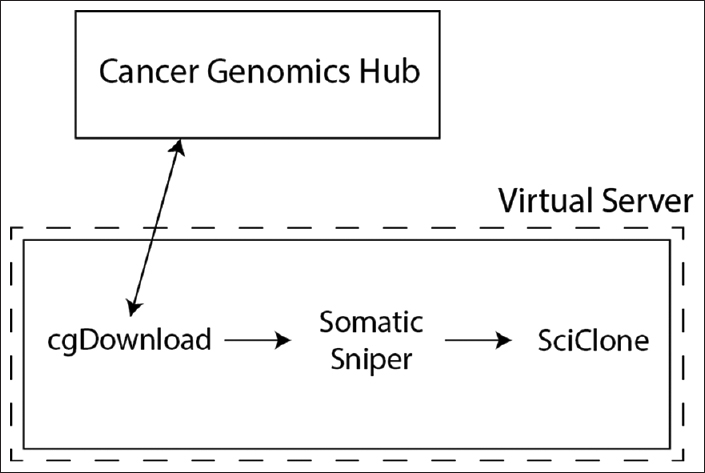 | 705 × 473 (89 KB) | Shawndouglas (talk | contribs) |
You cannot overwrite this file.
File usage
The following 2 pages use this file:









