File:Fig4 Durant JofPathInfo2020 11.jpg
Original file (1,210 × 367 pixels, file size: 184 KB, MIME type: image/jpeg)
Summary
| Description |
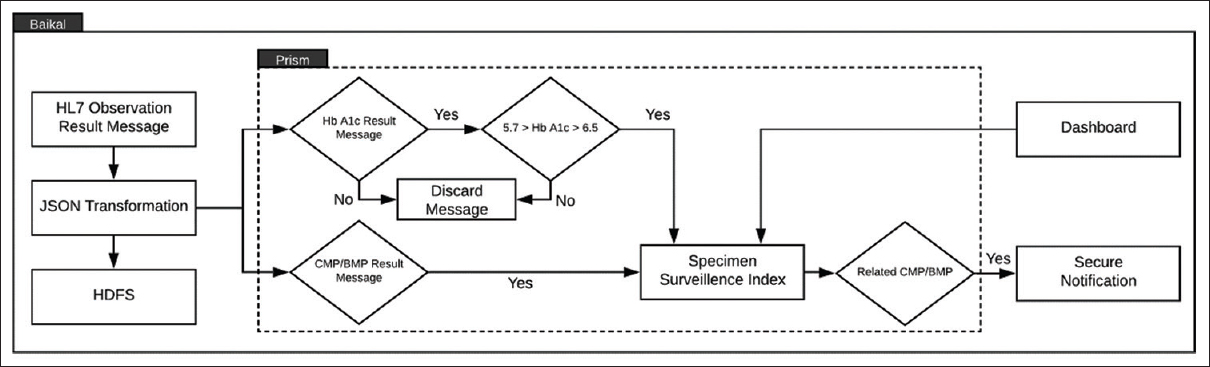
Fig. 4 Laboratory result processing diagram for diabetic biomarker monitoring. Incoming HL7 observations are transformed to denormalized JavaScript Object Notation documents and stored to HDFS. The Prism dataflow ingests streaming JavaScript Object Notation result records and filters hemoglobin A1C results in the “Normal” (<5.7) and “Diabetic” (>6.5) cohorts to the Prism Specimen Surveillance Index in Elastic. Results from CMP/BMP panels (light-green top specimens) are sent to the Prism index. Secure notifications are sent for A1C specimen IDs with related light-green specimen info. BMP: Basic Metabolic Panel, CMP: Comprehensive Metabolic Panel, HbA1c: Hemoglobin A1c; HDFS: Hadoop-Distributed File System, HL7: Health-Level 7, JSON: Java Script Object Notation |
|---|---|
| Source |
Durant, T.J.S.; Gong, G.; Price, N.; Schilz, W.L. (2020). "Bridging the collaboration gap: Real-time identification of clinical specimens for biomedical research". Journal of Pathology Informatics 11: 14. doi:10.4103/jpi.jpi_15_20. |
| Date |
2020 |
| Author |
Durant, T.J.S.; Gong, G.; Price, N.; Schilz, W.L. |
| Permission (Reusing this file) |
Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License |
| Other versions |
Licensing
|
|
This work is licensed under the Creative Commons Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. |
File history
Click on a date/time to view the file as it appeared at that time.
| Date/Time | Thumbnail | Dimensions | User | Comment | |
|---|---|---|---|---|---|
| current | 17:08, 2 June 2020 | 1,210 × 367 (184 KB) | Shawndouglas (talk | contribs) |
You cannot overwrite this file.
File usage
The following page uses this file:









