Journal:Building a newborn screening information management system from theory to practice
| Full article title | Building a newborn screening information management system from theory to practice |
|---|---|
| Journal | International Journal of Neonatal Screening |
| Author(s) | Pluscauskas, Michael; Henderson, Matthew; Milburn, Jennifer; Chakraborty, Pranesh |
| Author affiliation(s) | Children’s Hospital of Eastern Ontario, University of Ottawa |
| Primary contact | Email: mplus at cheo dot on dot ca |
| Year published | 2019 |
| Volume and issue | 5 (1) |
| Page(s) | 9 |
| DOI | 10.3390/ijns5010009 |
| ISSN | 2409-515X |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.mdpi.com/2409-515X/5/1/9/htm |
| Download | https://www.mdpi.com/2409-515X/5/1/9/pdf (PDF) |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
Information management systems are the central process management and communication hub for many newborn screening programs. In late 2014, Newborn Screening Ontario (NSO) undertook an end-to-end assessment of its information management needs, which resulted in a project to develop a flexible information systems (IS) ecosystem and related process changes. This enabled NSO to better manage its current and future workflow and communication needs. An idealized vision of a screening information management system (SIMS) was developed that was refined into enterprise and functional architectures. This was followed by the development of technical specifications, user requirements, and procurement. In undertaking a holistic full product lifecycle redesign approach, a number of change management challenges were faced by NSO across the entire program. Strong leadership support and full program engagement were key for overall project success. It is anticipated that improvements in program flexibility and the ability to innovate will outweigh the efforts and costs.
Keywords: newborn screening, neonatal screening, laboratory information management system, laboratory information system, LIMS, LIS, screening information management
Introduction
Dating back to the early days of computers, a variety of software programs have been utilized to automate numerous laboratory workflows, processes, and related activities. In newborn screening labs, a screening information management system (SIMS)—a set of integrated software components which may or may not be from a single vendor that can be used to automate screening workflows—has become a useful tool for automation. In addition, a SIMS can be used to manage other aspects of overall laboratory management and programmatic aspects, such as case reporting and follow-up.
Population-based newborn screening began appearing in many North American and European countries over 50 years ago. Due to the low number of tests per patient, many programs were initially able to handle their testing and reporting functions using manual workflows, paper-based lab logs, and patient reports. Labs initially employed simple computer functions such as using word processing to create mailing lists/labels and basic reports. Eventually, usage evolved into managing basic lab workflows and quality metrics using standard software such as spreadsheets and databases.
In the late 1990s and early 2000s, the shift to complex equipment such as tandem mass spectrometers (MS/MS), that could test for dozens of target diseases and related follow-ups, necessitated the development of a more complex information technology infrastructure to manage newborn screening programs. This shift required the development of new, often integrated, software programs to manage the new challenges that were created by these changes. Also, a number of new target disorders required very quick turnaround times (TATs) in order to locate infants who were at risk of early decompensation that could cause severe morbidity or even death. This meant that neonatal screening labs required information systems that could process large batches of results quickly in order to ensure that infants who tested positive for these disorders could be identified in a timely manner.
The current shift into new paradigms for newborn screening, including molecular (DNA-based) screening, is pushing the limits of the original software architectural approaches of these integrated packages. The ability of decision support tools such as Collaborative Laboratory Integrated Reports (CLIR)[1] to be linked, potentially in real time via application program interfaces (APIs)[2], also stretches the limits of current SIMS functionality. In addition, the potential of new point-of-care technologies and the possibility of screening for certain time-critical disorders prenatally have stretched these new paradigms even further. As these needs evolve, a more comprehensive information ecosystem approach to newborn screening SIMS architectures will be required to support the expanding needs of newborn screening.
In their paper “The Ideal Laboratory Information System,” Sepulveda and Young[3] described a set of processes and technology modules that they considered key for the development of an “ideal” comprehensive clinical laboratory information system (LIS). An approach to developing an “ideal” SIMS in a newborn screening context is described in this paper. The experiences of Newborn Screening Ontario (NSO) are used to illustrate the various technical and administrative approaches that are involved in undertaking such a project. The potential benefits, impacts, and risks of taking this approach are discussed and key lessons are highlighted.
Theory
Outgrowing NSO's current SIMS
NSO is located in the Canadian province of Ontario. Canada has a publicly funded health care system that is managed by its thirteen provinces and territories. Ontario is Canada’s largest province, with a population of approximately 14 million people and an annual public healthcare budget of over $CDN 50 billion in 2017. NSO is a fully integrated program that manages all aspects of newborn screening and follow-up activities for children born in the province of Ontario (approximately 140,000 births per year). With the expansion of provincial newborn screening in 2006, the lab and program management was moved from the provincial public health branch to the Children's Hospital of Eastern Ontario (CHEO), a leading academic pediatric research hospital that is located in the city of Ottawa. NSO receives ongoing annual base funding from the province as well as project-based funding to implement new programs as needed.
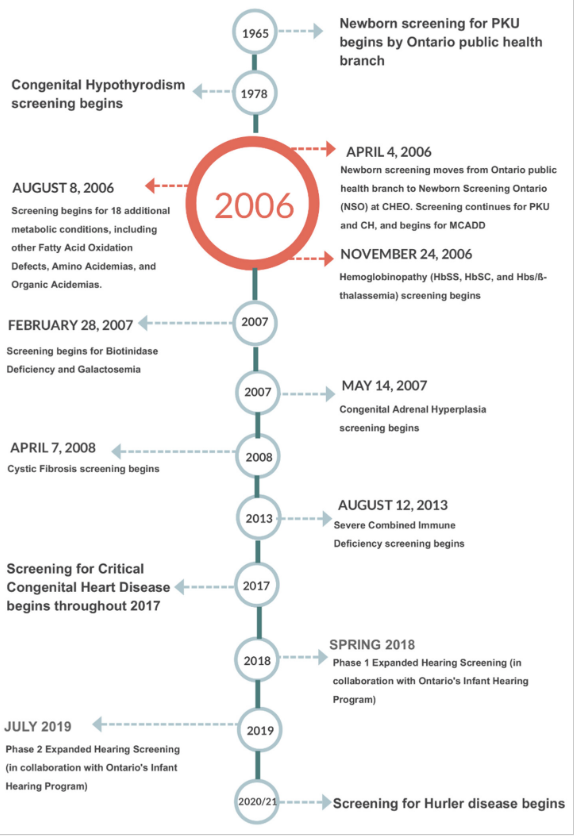
NSO performs blood-spot-based screening for metabolic and endocrine disorders as well as for hemoglobinopathies, cystic fibrosis (CF), and severe combined immune deficiencies (SCID). Multiple tier testing strategies are also used for a number of these diseases. In addition, NSO offers diagnostic and monitoring testing for many of the aforementioned disorders. Further, NSO coordinates and administers the provincial critical congenital heart disease (CCHD) point-of-care screening program.[4] NSO is also working with Ontario’s Infant Hearing Program (IHP) to phase in blood spot testing for hearing loss risk factors including cytomegalovirus (CMV) DNA and certain key genetic risk factors.[5] Figure 1 shows a timeline of newborn screening in Ontario from the beginning of province-wide screening for phenylketonuria (PKU) to the near future.
|
Like many other newborn screening programs, NSO coordinates and manages programmatic elements in addition to testing. These include pre-screening education, distribution of collection cards and communications, case management for special screening circumstances such as transfused or premature babies, short term follow-up of screen-positive babies, and analysis/reporting of key performance indicators. Finally, NSO has an academic mandate and coordinates regular meetings of treatment center physicians and health care providers, performs research, and participates in research collaborations, including work that is aimed at understanding the long-term outcomes of screening. It therefore requires an information infrastructure to support these diverse yet closely interrelated set of screening activities.
Newborn screening is a rapidly evolving field. An illustration of the rate of change in the last 15 years can be found in the adoption of the Recommended Universal Screening Panel (RUSP)[6] in the United States. In 2002, the American College of Medical Genetics reviewed 81 conditions and placed 29 of them in a core screening panel, which made up the original RUSP. At that time, the majority of U.S. states screened for only six disorders. Today, all states screen for at least 29 conditions.[7] The RUSP was, and continues to be, influential internationally.
A broader screening panel resulted in more test results per child screened, the need for novel analytical techniques, and the need for second/multiple tier testing to improve specificity. Like many other newborn screening labs, the NSO laboratory uses immunoassays, enzyme assays, chromatography, liquid chromatography–mass spectrometry (LC-MS/MS), flow injection analysis tandem mass spectrometry (FIA-MS/MS), quantitative real-time PCR (qPCR), and primer extension and next generation sequencing techniques for screening. Despite the breadth of techniques used, it is sometimes necessary to reflex first-tier screen-positive samples to second-tier testing methods in order to achieve an acceptable positive predictive value for a positive screening result. An example of tiered testing strategies and complex screening logic is congenital adrenal hyperplasia (CAH). First tier CAH screening relies on the measurement of 17-hydroxyprogesterone. The results are interpreted based on gestational age and birth-weight-specific screening thresholds. A panel of steroids is measured by LC-MS/MS in first-tier positive samples, and another set of screening logic is applied to this panel of results.
As the complexity of testing in newborn screening has increased, so has the capacity and expertise in the laboratory. As a result, newborn screening labs are well equipped to provide follow-up diagnostic and monitoring tests for screen-positive newborns and those affected by target diseases. In the case of NSO, we offer monitoring for patients with phenylketonuria, tyrosinemia, and glutaric aciduria type 1, along with molecular DNA testing for all disease targets of newborn screening. Finally, the pace of change in newborn screening requires that labs continue to develop and implement new screening, monitoring, and diagnostic approaches on an ongoing basis. NSO engages in small- to large-scale research studies that require data to be rapidly and readily available, including access to program, pre-analytical, testing, and follow-up data.
A single LIS solution for overall lab and program management was procured by NSO when it was established in 2006. This system was critical to the initial launch and subsequent development of the program, as it provided “tightly-coupled” support for the newborn screening laboratory information flows between equipment, quality control (QC), and case management. This approach worked well for ensuring the efficient management of lab workflows’ timely movement of information to follow-up teams, especially in relation to the standard blood spot testing for metabolic disorders. As the complexity of NSO’s mandate increased, it was determined that a broader approach was required to allow for the development of new areas. These areas included the expansion of molecular screening, diagnostic testing, complex screening algorithms, point-of-care screening, short- and long-term follow-up, and sample lifecycle management (see below). As noted above, it was determined that a broader newborn screening information ecosystem needed to be developed.
Designing an "ideal" SIMS for NSO
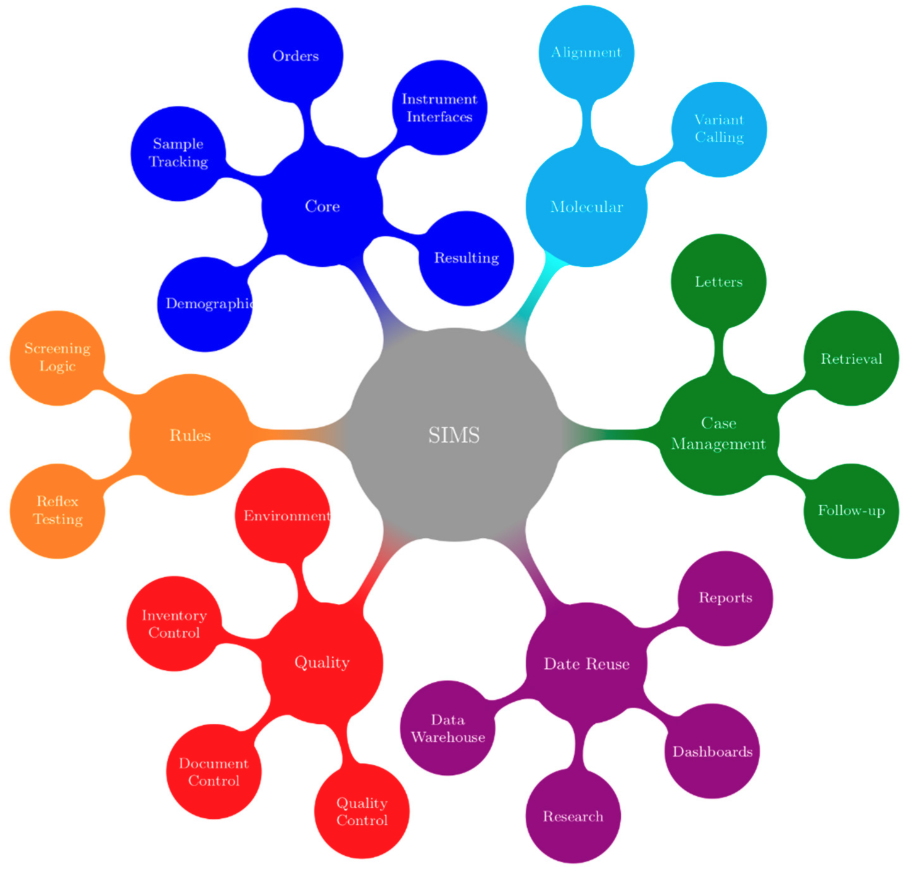
In their book, McCudden and Henderson noted “the present and future of health care relies on electronic information.”[8] The complexities that are required to capture complex demographics, manage high sample volumes in a timely manner, and integrate multiple test platforms have strained many programs’ abilities to move forward with these innovations. Newborn screening programs require a SIMS that is capable of managing all data coming into and going out of the program. There must also be facilities to customize the SIMS to the needs of the program. The functions of an ideal SIMS for newborn screening programs are outlined in Figure 2.
|
Screening for CF provides an illustrative example of the requirements of a SIMS; colors in the text are a reference to the concepts/functions that are illustrated in Figure 2. Generally, the first point at which information for a sample enters the SIMS is when samples arrive in the lab and demographics are entered or retrieved from hospital or jurisdictional registries (blue). Samples are punched for analysis (blue) in parallel with demographic entry to reduce turnaround time. The status of all samples from time of receipt in the lab to generation of positive or negative mailers is tracked using real-time dashboards that are located in the lab and administrative areas (purple). In many screening labs, an IRT-DNA CF screening strategy is used, with immunoreactivity trypsinogen (IRT) being used as a first-tier biomarker (blue). Testing is performed in accordance with relevant procedures (red). After review and acceptance of quality control results (red), a sample with elevated IRT will generate a request (orange) for CFTR genotyping (cyan). If screened positive for CF (orange), a risk letter will be generated (green). The risk letter incorporates both the IRT (blue) and CF genotype (cyan) results. Many jurisdictions have also implemented or are considering implementing third-tier sequencing that will further add to the complexity of this test. The program will refer the screen-positive infant to the appropriate care provider and this referral will be documented (green). The NBS program will be notified when the newborn is retrieved and this will be documented (green). Diagnostic testing will be performed and the NBS program will be informed of the final diagnosis (green). Periodically, the program will evaluate CF screening performance. This evaluation requires information from all aspects of the screening program (purple) such as, however not limited to, test turnaround time (blue), time to referral and retrieval (green), final or working diagnosis, and positive and negative predictive value (green). Babies and families with CF, CF variants, or false positive results may be invited to participate in further program evaluation and research, and the SIMS must facilitate this (green).
While not exhaustive, the CF screen-positive scenario provides an overview of the requirements of a SIMS for NBS. Not described above is the need to make changes to the system as disorders, methods, tests, and screening logic changes. Once changes are made, an approach to testing the system to ensure that it functions as intended will be required.
In order to realize this vision, NSO launched a large-scale project in late 2014 to develop a comprehensive service oriented architecture (SOA)[9] solution type in which information can flow seamlessly through the key areas of the program. The solution consisted of seven key functional modules (services) that, when combined, can achieve this vision, including:
1. Patient record management: This module is expected to handle most of the pre-analytical aspects of NSO. The module will be used to receive samples in the laboratory and to enter demographic data for these samples. It consists of a number of processes including but not limited to sample reception, demographic/clinical indication data entry and validation, test ordering (batch and custom), linking multiple samples to a patient; triggering workflows for samples that are unsatisfactory for testing, and ensuring electronic transmission receipt of key information (e.g., demographics in, results out) via HL7 and related protocols.
2. Laboratory information system/quality control: These modules will be expected to handle most of the analytical and QC data, including workflows and dataflows within the NSO laboratory environment.
3. Clinical/medical review: The review and releasing of results is a critical component of laboratory information workflow between the technologists and medical/scientific staff. This module is designed to apply pre-programmed logic to distill critical lab information in order to produce actionable results in an efficient manner. The clinical/medical review module will consist of a configurable rules-based system that integrates information from multiple data sources, applies disorder logic, and streamlines workflows to drive decisions. It will also include a rules-based expert system, including a web-based graphical user interface (GUI) to support the review and reporting functions, an administrative interface for creating and managing rules, and a user-configurable knowledge base that is “human readable.” In its first iteration, this functionality will be embedded in the core SIMS. The possibility of using an independent rules engine service that can be made available to multiple systems in the longer term will also be explored.
4. Case management: This module will be expected to handle most of the post-analytical aspects of NSO, including follow-up with submitters and treatment centers.
5. Sample lifecycle management: The need to track blood collection cards throughout their lifecycle, from distribution through transport to storage, is a key and sometimes overlooked process within newborn screening program management. A local vendor has worked with NSO to develop a system that helps manage sample card inventory management, track the transport of samples and card usage, and monitor expiry dates for filter paper; the vendor is also working towards tracking in-lab samples, off-site storage, and destruction.
6. Reporting and analytics: In order to support the data intensive nature of newborn screening results, NSO has developed a data warehouse to enable user-controlled data access and to manage automated and ad-hoc reporting on all types of NSO data.
7. Decision support: Third-party products can be linked in to provide decision support to key decision makers to assist in timely, effective decision making.
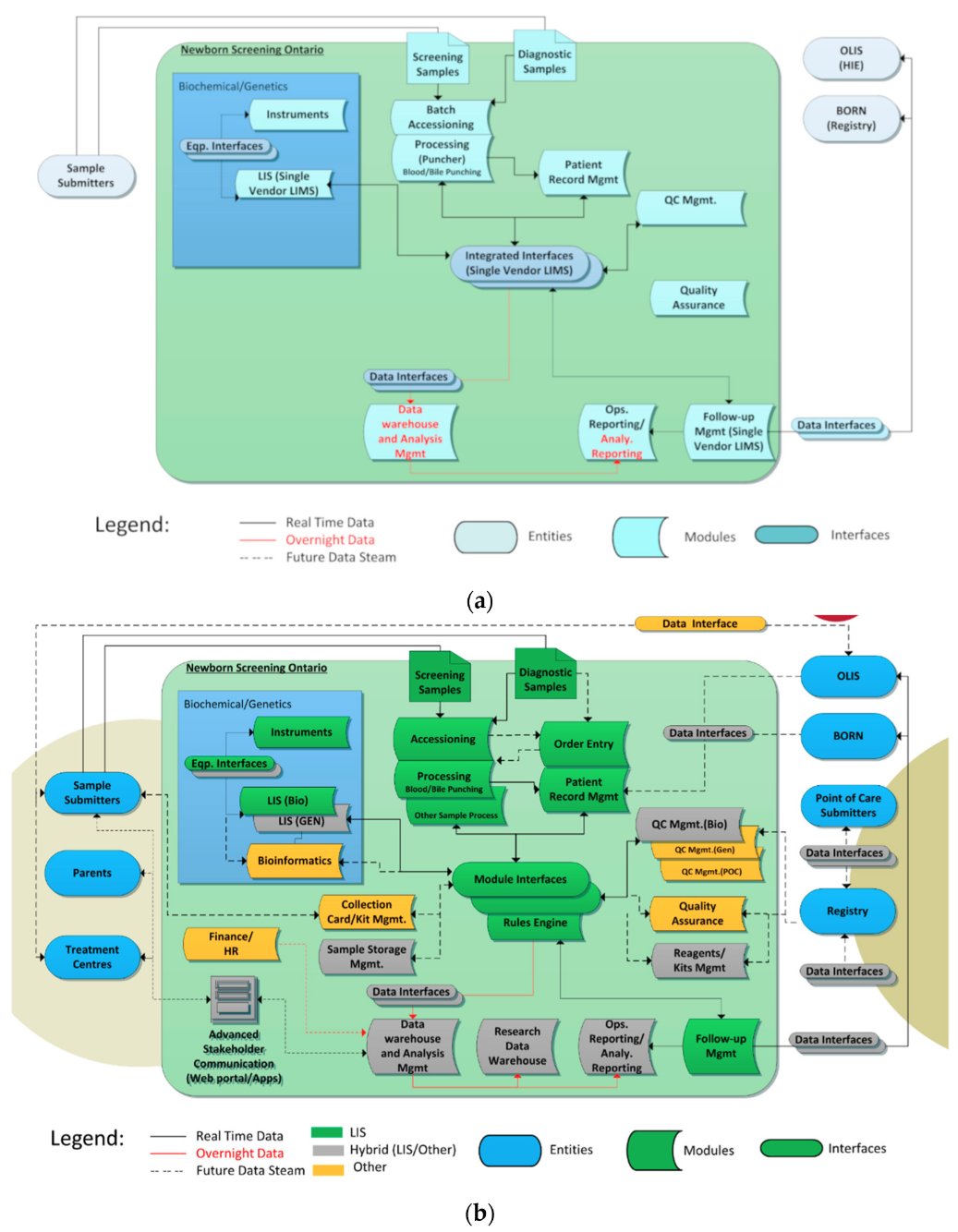
These functional modules were combined to produce an enterprise architecture to help guide the project. Figure 3a shows the “tightly coupled” enterprise architecture of the original NSO information system infrastructure, and Figure 3b shows the desired final state.
|
The NSO SIMS components are shown within the large blue box including instruments/interfaces, LISs, and related systems such as data warehouses. The ovals outside the large box show NSO’s data consumers (entities), which includes key groups such as hospitals who submit samples (submitters) and treatment centers who retrieve screen-positive and parents. They also show data holding entities such as health information exchanges (HIEs) and data registries. Dataflows are shown via directional arrows.
In Figure 3a, the blue color represents the fact that all functions were handled by a single “tightly coupled” LIS. The new enterprise architecture is more “services” focused with different systems playing different roles. The colors in Figure 3b call out what components are responsible for each function in the new NSO data architecture, where green is the main LIS (OMNILab), gray represents a hybrid of the main LIS and other in-house systems, and orange represents other off-the-shelf components.
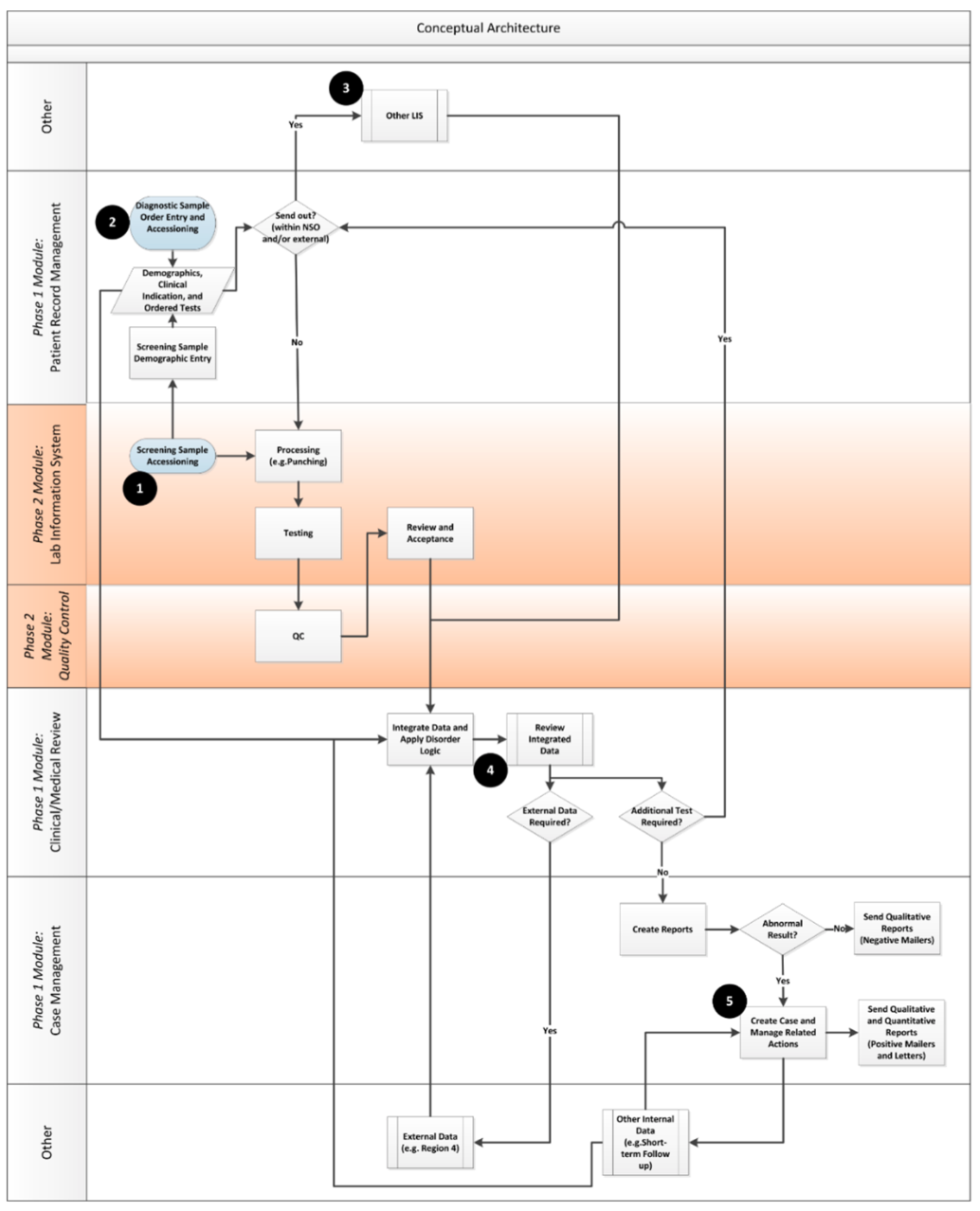
Once the enterprise view of the desired enterprise architecture was developed, the team moved on to ensure that key functional considerations for the system were taken into account by developing a functional architecture. An analogy of the flow from enterprise architecture to a functional one is similar to the link between a building’s architecture and building plans. The enterprise architecture provides a high-level, idealized view of what can be accomplished across the organization, whereas the functional views (with related user requirements) provide detail about what needs to be built. Figure 4 shows the key conceptual information flows that were developed by the team to guide the project. The modules and phases of implementation are called out on the right side of the diagram.
|
References
- ↑ "About CLIR". Mayo Foundation for Medical Education and Research. https://clir.mayo.edu/Home/About. Retrieved 26 November 2018.
- ↑ Pantanowitz, L.; Henricks, W.H.; Beckwith, B.A. (2007). "Medical laboratory informatics". Clinics in Laboratory Medicine 27 (4): 823–43. doi:10.1016/j.cll.2007.07.011. PMID 17950900.
- ↑ Sepulveda, J.L;. Young, D.S. (2013). "The ideal laboratory information system". pp. 1129–40. doi:10.5858/arpa.2012-0362-RA. PMID 23216205.
- ↑ "CCHD Screening". Newborn Screening Ontario. https://www.newbornscreening.on.ca/en/health-care-providers/submitters/cchd-screening-implementation. Retrieved 26 November 2018.
- ↑ "Expanded Hearing Screenings - Overview". Newborn Screening Ontario. https://www.newbornscreening.on.ca/en/page/overview. Retrieved 26 November 2018.
- ↑ "Recommended Uniform Screening Panel". Health Resources & Services Administration. July 2018. https://www.hrsa.gov/advisory-committees/heritable-disorders/rusp/index.html. Retrieved 26 November 2018.
- ↑ Secretary’s Advisory Committee on Heritable Disorders in Newborns and Children (2011). "2011 Annual Report to Congress" (PDF). Health Resources & Services Administration. https://www.hrsa.gov/sites/default/files/hrsa/advisory-committees/heritable-disorders/reports-recommendations/reports/2011-annual-report.pdf. Retrieved 20 February 2018.
- ↑ McCudden, C.R.; Henderson, M.P.A. (2016). "Laboratory Information Systems". In Clarke, W.. Contemporary Practice in Clinical Chemistry (3rd ed.). pp. 263–76. ISBN 9781594251894.
- ↑ Huhns, M.S.; Singh, M.P. (2005). "Service-oriented computing: key concepts and principles". IEEE Internet Computing 9 (1): 75–81. doi:10.1109/MIC.2005.21.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In several cases the PubMed ID was missing and was added to make the reference more useful.