Difference between revisions of "Journal:Characterizing and managing missing structured data in electronic health records: Data analysis"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) (Saving and adding more.) |
||
| Line 62: | Line 62: | ||
When data are missing in a manner completely unrelated to both the observed and unobserved values, they are considered to be missing completely at random (MCAR).<ref name="WellsStrat13">{{cite journal |title=Strategies for handling missing data in electronic health record derived data |journal=EGEMS |author=Wells, B.J.; Chagin, K.M.; Nowacki, A.S.; Kattan, M.W. |volume=1 |issue=3 |pages=1035 |doi=10.13063/2327-9214.1035 |pmid=25848578 |pmc=PMC4371484}}</ref><ref name="BounthavongApproach15">{{cite journal |title=Approach to addressing missing data for electronic medical records and pharmacy claims data research |journal=Pharmacotherapy |author=Bounthavong, M.; Watanabe, J.H.; Sullivan, K.M. |volume=35 |issue=4 |pages=380–7 |doi=10.1002/phar.1569 |pmid=25884526}}</ref> When data are MCAR, the observed data represent a random sample of the population, but this is rarely encountered in practice. Conversely, data missing not at random (MNAR) refers to a situation where the probability of observing a data point depends on the value of that data point.<ref name="BhaskaranWhat14">{{cite journal |title=What is the difference between missing completely at random and missing at random? |journal=International Journal of Epidemiology |author=Bhaskaran, K.; Smeeth, L. |volume=43 |issue=4 |pages=1336-9 |doi=10.1093/ije/dyu080 |pmid=24706730 |pmc=PMC4121561}}</ref> In this case, the mechanism responsible for the missing data is biased and should not be considered ignorable.<ref name="RubinInference76">{{cite journal |title=Inference and missing data |journal=Biometrika |author=Rubin, D.B. |volume=63 |issue=3 |pages=581–592 |doi=10.1093/biomet/63.3.581}}</ref> For example, rheumatoid factor is an antibody detectable in blood, and the concentration of this antibody is correlated with the presence and severity of rheumatoid arthritis. This test is typically performed only for patients with some indication of rheumatoid arthritis. Thus, patients with high rheumatoid factor levels are more likely to have rheumatoid factor measures. | When data are missing in a manner completely unrelated to both the observed and unobserved values, they are considered to be missing completely at random (MCAR).<ref name="WellsStrat13">{{cite journal |title=Strategies for handling missing data in electronic health record derived data |journal=EGEMS |author=Wells, B.J.; Chagin, K.M.; Nowacki, A.S.; Kattan, M.W. |volume=1 |issue=3 |pages=1035 |doi=10.13063/2327-9214.1035 |pmid=25848578 |pmc=PMC4371484}}</ref><ref name="BounthavongApproach15">{{cite journal |title=Approach to addressing missing data for electronic medical records and pharmacy claims data research |journal=Pharmacotherapy |author=Bounthavong, M.; Watanabe, J.H.; Sullivan, K.M. |volume=35 |issue=4 |pages=380–7 |doi=10.1002/phar.1569 |pmid=25884526}}</ref> When data are MCAR, the observed data represent a random sample of the population, but this is rarely encountered in practice. Conversely, data missing not at random (MNAR) refers to a situation where the probability of observing a data point depends on the value of that data point.<ref name="BhaskaranWhat14">{{cite journal |title=What is the difference between missing completely at random and missing at random? |journal=International Journal of Epidemiology |author=Bhaskaran, K.; Smeeth, L. |volume=43 |issue=4 |pages=1336-9 |doi=10.1093/ije/dyu080 |pmid=24706730 |pmc=PMC4121561}}</ref> In this case, the mechanism responsible for the missing data is biased and should not be considered ignorable.<ref name="RubinInference76">{{cite journal |title=Inference and missing data |journal=Biometrika |author=Rubin, D.B. |volume=63 |issue=3 |pages=581–592 |doi=10.1093/biomet/63.3.581}}</ref> For example, rheumatoid factor is an antibody detectable in blood, and the concentration of this antibody is correlated with the presence and severity of rheumatoid arthritis. This test is typically performed only for patients with some indication of rheumatoid arthritis. Thus, patients with high rheumatoid factor levels are more likely to have rheumatoid factor measures. | ||
A more complicated scenario can arise when multiple variables are available. If the probability of observing a data point does not depend on the value of that data point, after conditioning on one or more additional variables, then that data point is said to be missing at random (MAR).<ref name="BhaskaranWhat14" /> For example, a variable, ''X'', may be MNAR if considered in isolation. However, if we observe another variable, ''Y'', that explains some of the variation in ''X'' such that, after conditioning on ''Y'', the probability of observing ''X'' is no longer related to its own value, then ''X'' is said to be MAR. In this way, ''Y'' can transform ''X'' from MNAR to MAR (Figure 1). We cannot prove that ''X'' is randomly sampled unless we measure some of the unobserved values, but strong correlations, the ability to explain missingness, and domain knowledge may provide evidence that the data are MAR. | |||
Imputation methods assume specific mechanisms of missingness, and assumption violations can lead to bias in the results of downstream analyses that can be difficult to predict.<ref name="JörnstenAMeta07">{{cite journal |title=A meta-data based method for DNA microarray imputation |journal=BMC Bioinformatics |author=Jörnsten, R.; Ouyang, M.; Wang, H.Y. |volume=8 |pages=109 |doi=10.1186/1471-2105-8-109 |pmid=17394658 |pmc=PMC1852325}}</ref><ref name="Beaulieu-JonesMissing17">{{cite journal |title=Missing data imputation in the electronic health record using deeply learned autoencoders |journal=Pacific Symposium on Biocomputing |author=Beaulieu-Jones, B.K.; Moore, J.H. |volume=22 |pages=207-218 |doi=10.1142/9789813207813_0021 |pmid=27896976 |pmc=PMC5144587}}</ref> Variances of imputed values are often underestimated, causing artificially low ''P'' values.<ref name="AllisonMissing02">{{cite book |title=Missing Data |author=Allison, P.D. |publisher=SAGE Publications |volume=136 |series=Quantitative Applications in the Social Sciences |year=2002 |isbn=9781412985079 |doi=10.4135/9781412985079}}</ref> Additionally, for data MNAR, the observed values have a different distribution from the missing values. To cope with this, a model can be specified to represent the missing data mechanism, but such models can be difficult to evaluate and may have a large impact on results. Great caution should be taken when handling missing data, particularly data that are MNAR. Most imputation methods assume that data are MAR or MCAR, but it is worth reiterating that these are all idealized states, and real data invariably fall somewhere in between (Figure 1). | |||
===Objective=== | |||
We aimed to provide a framework for characterizing and understanding the types of missing data present in the EHR. We also developed an open-source framework that other researchers can follow when dealing with missing data. | |||
==Methods== | |||
===Source code=== | |||
We provide the source code to reproduce this work in our repository on GitHub (GitHub, Inc.)<ref name="GHImputation">{{cite web |url=https://github.com/EpistasisLab/imputation |title=EpistasisLab/imputation |publisher=GitHub, Inc |accessdate=01 December 2017}}</ref> under a permissive open source license. In addition, we used continuous analysis<ref name="Beaulieu-JonesRepro17">{{cite journal |title=Reproducibility of computational workflows is automated using continuous analysis |journal=Nature Biotechnology |author=Beaulieu-Jones, B.K.; Greene, C.S. |volume=35 |issue=4 |pages=342-346 |doi=10.1038/nbt.3780 |pmid=28288103}}</ref> to generate Docker Hub (Docker Inc.) images matching the environment of the original analysis and to create intermediate results and logs. These artifacts are freely available.<ref name="DockerImputation">{{cite web |url=https://hub.docker.com/r/brettbj/ehr-imputation/ |title=brettbj/ehr-imputation |publisher=Docker, Inc |accessdate=02 December 2017}}</ref> | |||
===Electronic health record data processing=== | |||
All laboratory assays were mapped to Logical Observation Identifiers Names and Codes (LOINC). We restricted our analysis to outpatient laboratory results to minimize the effects of extreme results from inpatient and emergency department data. We used all laboratory results dated between August 8, 1996 and March 3, 2016, excluding codes for which less than 0.5% of patients had a result. The resulting dataset consisted of 669,212 individuals and 143 laboratory assays. | |||
We removed any laboratory results that were obtained prior to the patient’s 18th birthday or after their 90th. In cases where a date of death was present, we also removed laboratory results that were obtained within one year of death, as we found that the frequency of observations often spiked during this period and the values for certain laboratory tests were altered for patients near death. For each patient, a median date of observation was calculated based on their remaining laboratory results. We defined a temporal window of observation by removing any laboratory results recorded more than five years from the median date. We then calculated the median result of the remaining laboratory tests for each patient. As each variable had a different scale and many deviated from normality, we applied Box-Cox and Z-transformations to all variables. The final dataset used for all downstream analyses contained 602,366 patients and 146 variables (age, sex, body mass index [BMI], and 143 laboratory measures). | |||
===Variable selection=== | |||
We first ranked the laboratory measures by total amount of missingness, lowest to highest. At each rank, we calculated the percentage of complete cases for the set, including all lower-ranked measures. We also built a random forest classifier to predict the presence or absence of each variable. Based on these results and domain knowledge, we selected 28 variables that provided a reasonable trade-off between quantity and completeness and that we deemed to be largely MAR. | |||
==References== | ==References== | ||
Revision as of 18:43, 6 March 2018
| Full article title | Characterizing and managing missing structured data in electronic health records: Data analysis |
|---|---|
| Journal | JMIR Medical Informatics |
| Author(s) | Beaulieu-Jones, Brett K.; Lavage, Daniel R.; Snyder, John W.; Moore, Jason H.; Pendergrass, Sarah A.; Bauer, Christopher R. |
| Author affiliation(s) | University of Pennsylvania, Geisinger |
| Primary contact | Email: cbauer at geisinger dot edu |
| Year published | 2018 |
| Volume and issue | 6 (1) |
| Page(s) | e11 |
| DOI | 10.2196/medinform.8960 |
| ISSN | 2291-9694 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | http://medinform.jmir.org/2018/1/e11/ |
| Download | http://medinform.jmir.org/2018/1/e11/pdf (PDF) |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
Background: Missing data is a challenge for all studies; however, this is especially true for electronic health record (EHR)-based analyses. Failure to appropriately consider missing data can lead to biased results. While there has been extensive theoretical work on imputation, and many sophisticated methods are now available, it remains quite challenging for researchers to implement these methods appropriately. Here, we provide detailed procedures for when and how to conduct imputation of EHR laboratory results.
Objective: The objective of this study was to demonstrate how the mechanism of "missingness" can be assessed, evaluate the performance of a variety of imputation methods, and describe some of the most frequent problems that can be encountered.
Methods: We analyzed clinical laboratory measures from 602,366 patients in the EHR of Geisinger Health System in Pennsylvania, USA. Using these data, we constructed a representative set of complete cases and assessed the performance of 12 different imputation methods for missing data that was simulated based on four mechanisms of missingness (missing completely at random, missing not at random, missing at random, and real data modelling).
Results: Our results showed that several methods, including variations of multivariate imputation by chained equations (MICE) and softImpute, consistently imputed missing values with low error; however, only a subset of the MICE methods was suitable for multiple imputation.
Conclusions: The analyses we describe provide an outline of considerations for dealing with missing EHR data, steps that researchers can perform to characterize missingness within their own data, and an evaluation of methods that can be applied to impute clinical data. While the performance of methods may vary between datasets, the process we describe can be generalized to the majority of structured data types that exist in EHRs, and all of our methods and code are publicly available.
Keywords: imputation, missing data, clinical laboratory test results, electronic health records
Introduction
Justification
Missing data present a challenge to researchers in many fields, and this challenge is growing as datasets increase in size and scope. This is especially problematic for electronic health records (EHRs), where missing values frequently outnumber observed values. EHRs were designed to record and improve patient care and streamline billing rather than act as resources for research[1]; thus, there are significant challenges to using these data to gain a better understanding of human health. As EHR data become increasingly used as a source of phenotypic information for biomedical research[2], it is crucial to develop strategies for coping with missing data.
Clinical laboratory assay results are a particularly rich data source within the EHR, but they also tend to have large amounts of missing data. These data may be missing for many different reasons. Some tests are used for routine screening, but screening may be biased. Other tests are only conducted if they are clinically relevant to very specific ailments. Patients may also receive care at multiple health care systems, resulting in information gaps at each institution. Age, sex, socioeconomic status, access to care, and medical conditions can all affect how comprehensive the data are for a given patient. Accounting for the mechanisms that cause data to be missing is critical, since failure to do so can lead to biased conclusions.
Background
Aside from the uncertainty associated with a variable that is not observed, many analytical methods, such as regression or principal components analysis, are designed to operate only on a complete dataset. The easiest way to implement these procedures is to remove variables with missing values or remove individuals with missing values. Eliminating variables is justifiable in many situations, especially if a given variable has a large proportion of missing values, but doing so may restrict the scope and power of a study. Removing individuals with missing data is another option known as complete-case analysis. This is generally not recommended unless the fraction of individuals that will be removed is small enough to be considered trivial, or there is good reason to believe that the absence of a value is due to random chance. If there are systematic differences between individuals with and without observations, complete-case analysis will be biased.
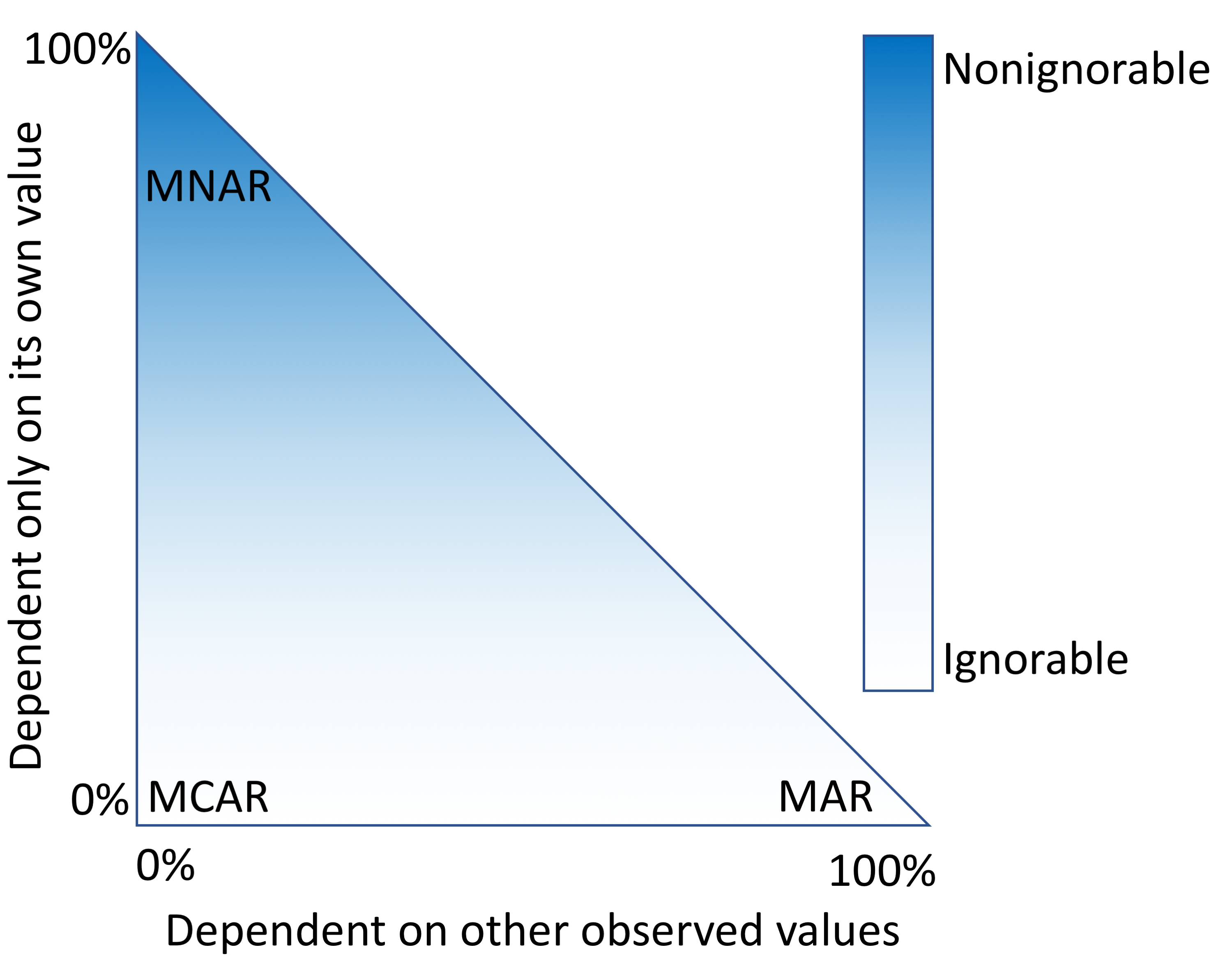
An alternative approach is to fill in the fields that are missing data with estimates. This process, called imputation, requires a model that makes assumptions about why only some values were observed. "Missingness" mechanisms fall somewhere in a spectrum between three scenarios (Figure 1).
|
When data are missing in a manner completely unrelated to both the observed and unobserved values, they are considered to be missing completely at random (MCAR).[3][4] When data are MCAR, the observed data represent a random sample of the population, but this is rarely encountered in practice. Conversely, data missing not at random (MNAR) refers to a situation where the probability of observing a data point depends on the value of that data point.[5] In this case, the mechanism responsible for the missing data is biased and should not be considered ignorable.[6] For example, rheumatoid factor is an antibody detectable in blood, and the concentration of this antibody is correlated with the presence and severity of rheumatoid arthritis. This test is typically performed only for patients with some indication of rheumatoid arthritis. Thus, patients with high rheumatoid factor levels are more likely to have rheumatoid factor measures.
A more complicated scenario can arise when multiple variables are available. If the probability of observing a data point does not depend on the value of that data point, after conditioning on one or more additional variables, then that data point is said to be missing at random (MAR).[5] For example, a variable, X, may be MNAR if considered in isolation. However, if we observe another variable, Y, that explains some of the variation in X such that, after conditioning on Y, the probability of observing X is no longer related to its own value, then X is said to be MAR. In this way, Y can transform X from MNAR to MAR (Figure 1). We cannot prove that X is randomly sampled unless we measure some of the unobserved values, but strong correlations, the ability to explain missingness, and domain knowledge may provide evidence that the data are MAR.
Imputation methods assume specific mechanisms of missingness, and assumption violations can lead to bias in the results of downstream analyses that can be difficult to predict.[7][8] Variances of imputed values are often underestimated, causing artificially low P values.[9] Additionally, for data MNAR, the observed values have a different distribution from the missing values. To cope with this, a model can be specified to represent the missing data mechanism, but such models can be difficult to evaluate and may have a large impact on results. Great caution should be taken when handling missing data, particularly data that are MNAR. Most imputation methods assume that data are MAR or MCAR, but it is worth reiterating that these are all idealized states, and real data invariably fall somewhere in between (Figure 1).
Objective
We aimed to provide a framework for characterizing and understanding the types of missing data present in the EHR. We also developed an open-source framework that other researchers can follow when dealing with missing data.
Methods
Source code
We provide the source code to reproduce this work in our repository on GitHub (GitHub, Inc.)[10] under a permissive open source license. In addition, we used continuous analysis[11] to generate Docker Hub (Docker Inc.) images matching the environment of the original analysis and to create intermediate results and logs. These artifacts are freely available.[12]
Electronic health record data processing
All laboratory assays were mapped to Logical Observation Identifiers Names and Codes (LOINC). We restricted our analysis to outpatient laboratory results to minimize the effects of extreme results from inpatient and emergency department data. We used all laboratory results dated between August 8, 1996 and March 3, 2016, excluding codes for which less than 0.5% of patients had a result. The resulting dataset consisted of 669,212 individuals and 143 laboratory assays.
We removed any laboratory results that were obtained prior to the patient’s 18th birthday or after their 90th. In cases where a date of death was present, we also removed laboratory results that were obtained within one year of death, as we found that the frequency of observations often spiked during this period and the values for certain laboratory tests were altered for patients near death. For each patient, a median date of observation was calculated based on their remaining laboratory results. We defined a temporal window of observation by removing any laboratory results recorded more than five years from the median date. We then calculated the median result of the remaining laboratory tests for each patient. As each variable had a different scale and many deviated from normality, we applied Box-Cox and Z-transformations to all variables. The final dataset used for all downstream analyses contained 602,366 patients and 146 variables (age, sex, body mass index [BMI], and 143 laboratory measures).
Variable selection
We first ranked the laboratory measures by total amount of missingness, lowest to highest. At each rank, we calculated the percentage of complete cases for the set, including all lower-ranked measures. We also built a random forest classifier to predict the presence or absence of each variable. Based on these results and domain knowledge, we selected 28 variables that provided a reasonable trade-off between quantity and completeness and that we deemed to be largely MAR.
References
- ↑ Steinbrook, R.. "Health Care and the American Recovery and Reinvestment Act". New England Journal of Medicine 360 (11): 1057–1060. doi:10.1056/NEJMp0900665. PMID 19224738.
- ↑ Flintoft, L.. "Disease genetics: Phenome-wide association studies go large". Nature Reviews Genetics 15 (1): 2. doi:10.1038/nrg3637. PMID 24322724.
- ↑ Wells, B.J.; Chagin, K.M.; Nowacki, A.S.; Kattan, M.W.. "Strategies for handling missing data in electronic health record derived data". EGEMS 1 (3): 1035. doi:10.13063/2327-9214.1035. PMC PMC4371484. PMID 25848578. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4371484.
- ↑ Bounthavong, M.; Watanabe, J.H.; Sullivan, K.M.. "Approach to addressing missing data for electronic medical records and pharmacy claims data research". Pharmacotherapy 35 (4): 380–7. doi:10.1002/phar.1569. PMID 25884526.
- ↑ 5.0 5.1 Bhaskaran, K.; Smeeth, L.. "What is the difference between missing completely at random and missing at random?". International Journal of Epidemiology 43 (4): 1336-9. doi:10.1093/ije/dyu080. PMC PMC4121561. PMID 24706730. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4121561.
- ↑ Rubin, D.B.. "Inference and missing data". Biometrika 63 (3): 581–592. doi:10.1093/biomet/63.3.581.
- ↑ Jörnsten, R.; Ouyang, M.; Wang, H.Y.. "A meta-data based method for DNA microarray imputation". BMC Bioinformatics 8: 109. doi:10.1186/1471-2105-8-109. PMC PMC1852325. PMID 17394658. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1852325.
- ↑ Beaulieu-Jones, B.K.; Moore, J.H.. "Missing data imputation in the electronic health record using deeply learned autoencoders". Pacific Symposium on Biocomputing 22: 207-218. doi:10.1142/9789813207813_0021. PMC PMC5144587. PMID 27896976. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5144587.
- ↑ Allison, P.D. (2002). Missing Data. Quantitative Applications in the Social Sciences. 136. SAGE Publications. doi:10.4135/9781412985079. ISBN 9781412985079.
- ↑ "EpistasisLab/imputation". GitHub, Inc. https://github.com/EpistasisLab/imputation. Retrieved 01 December 2017.
- ↑ Beaulieu-Jones, B.K.; Greene, C.S.. "Reproducibility of computational workflows is automated using continuous analysis". Nature Biotechnology 35 (4): 342-346. doi:10.1038/nbt.3780. PMID 28288103.
- ↑ "brettbj/ehr-imputation". Docker, Inc. https://hub.docker.com/r/brettbj/ehr-imputation/. Retrieved 02 December 2017.
Notes
This presentation is faithful to the original, with only a few minor changes to grammar, spelling, and presentation, including the addition of PMCID and DOI when they were missing from the original reference.