Difference between revisions of "Journal:Electronic laboratory notebooks in a public–private partnership"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) (Saving and adding more.) |
||
| Line 85: | Line 85: | ||
In this article we describe the process of selecting and implementing an ELN in the context of the multisite PPP project TRANSLOCATION, comprising about 90 bench scientists in total. Furthermore we present the results from a survey evaluating the users’ experiences and the benefit for the project two years post-implementation. Based on our experiences, the specific needs in a PPP setting are summarized and lessons learned will be reviewed. As a result, we propose recommendations to assist future users avoiding pitfalls when selecting and implementing ELN software. | In this article we describe the process of selecting and implementing an ELN in the context of the multisite PPP project TRANSLOCATION, comprising about 90 bench scientists in total. Furthermore we present the results from a survey evaluating the users’ experiences and the benefit for the project two years post-implementation. Based on our experiences, the specific needs in a PPP setting are summarized and lessons learned will be reviewed. As a result, we propose recommendations to assist future users avoiding pitfalls when selecting and implementing ELN software. | ||
==Methods== | |||
===Selection and implementation of an ELN solution=== | |||
The IMI project call requested a high level of transparency enabling the sharing of data to serve as an example for future projects. The selected consortium TRANSLOCATION had a special demand for an ELN due to its structure — various labs and partners spread widely across Europe needed to report into one common repository — and due to the final goal, data was required to be stored and integrated into one central information hub, the ND4BB Information Centre. Fortunately, no legacy data had to be migrated into the ELN. | |||
The standard process for the introduction of new software follows a highly structured multi-phase procedure<ref name="NehmeElect06">{{cite book |chapter=9. Electronic Laboratory Notebooks |title=Computer Applications in Pharmaceutical Research and Development |author=Nehme, A.; Scoffin, R.A. |editor=Ekins, S.; Wang, B. |publisher=Wiley |year=2006 |pages=209–27 |isbn=9780471737797}}</ref><ref name="MilstedLabTrove13">{{cite journal |title=LabTrove: a lightweight, web based, laboratory "blog" as a route towards a marked up record of work in a bioscience research laboratory |journal=PLoS One |author=Milsted, A.J.; Hale, J.R.; Grey, J.G.; Neylon, C. |volume=8 |issue=7 |pages=e67460 |year=2013 |doi=10.1371/journal.pone.0067460 |pmid=23935832 |pmc=PMC3720848}}</ref>, as outlined in Fig. 2. | |||
[[File:Fig2 Vaas PeerJCompSci2016 2.jpg|500px]] | |||
{{clear}} | |||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| border="0" cellpadding="5" cellspacing="0" width="500px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 2.''' Schematic outline of stepwise procedure for the implementation of a new system<ref name="NehmeElect06" /></blockquote> | |||
|- | |||
|} | |||
|} | |||
For the first step, we had to manage a large and highly heterogeneous user group (Fig. 3) that would be using the ELN, scheduled for roll-out within six months after project launch. All personnel of the academic partners were requested to enter data into the same ELN, potentially leading to unmet individual user requirements, especially for novices and inexperienced users. | |||
[[File:Fig3 Vaas PeerJCompSci2016 2.jpg|500px]] | |||
{{clear}} | |||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| border="0" cellpadding="5" cellspacing="0" width="500px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 3.''' Technical and organizational challenge: schematic overview of paths for sharing research activity results within a public–private partnership on antimicrobial research</blockquote> | |||
|- | |||
|} | |||
|} | |||
As a compromise for step 1 (Fig. 2), we assembled a collection of user requirement specifications (URS) based on the experiences of one laboratory that had already implemented an ELN. We further selected a small group of super users based on their expertise in documentation processes, representing different wet laboratories and in silico environments. The resulting URS was reviewed by IT and business experts from academic as well as private organisations of the consortium. The final version of the URS is available as Supplemental File 1. | |||
In parallel, based on literature (Rubacha, Rattan & Hossel, 2011) and internet searches, presentations of widely used ELNs were evaluated to gain insight into state-of-the-art ELNs. This revealed a wide variety of functional and graphical user interface (GUI) implementations differing in complexity and costs. The continuum between simple out-of-the-box solutions and highly sophisticated and configurable ELNs with interfaces to state-of-the-art analytical tools were covered by the presentations. Notably, the requirements specified by super users also ranged from “easy to use” to “highly individually configurable.” Based on this information, it was clear that the ELN selected for this consortium would never ideally fit all user expectations. Furthermore, the exact number of users and configuration of user groups were unknown at the onset of the project. The most frequently or highest prioritized items of the collected user requirements are listed in Table 2. We divided the gathered requirements into ‘core’ meaning essential and ‘non-core’ meaning ‘nice to have, but not indispensable.’ Further, we list here only the items, which were mentioned by more than two super users from different groups. The full list of URS is available in Supplemental File 1. | |||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| class="wikitable" border="1" cellpadding="5" cellspacing="0" width="70%" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" colspan="2"|'''Table 2.''' Overview of user requirements organized as ‘core user requirements’ for essential items, and ‘non-core user requirements’ representing desirable features | |||
|- | |||
! style="padding-left:10px; padding-right:10px;"|Core user requirements | |||
! style="padding-left:10px; padding-right:10px;"|Non-core user requirements | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
• System set-up and implementation should be fast and simple<br /> | |||
• Access from different platforms should be possible: Windows, Linux, Mac OS<br /> | |||
• Low training requirements (for high level of acceptance<br /> | |||
• Hosted system with state-of-the-art security settings<br /> | |||
• Simple user management (only limited support by project members possible)<br /> | |||
• Suitable for both chemical (including e.g., drawings of molecules) and biological (including e.g., capture fluorescent images) experiments<br /> | |||
• Low costs, especially for long-term usage in the academic area<br /> | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| | |||
• Conform with Good Laboratory Practise<br /> | |||
• User management with dedicated access permissions (expectation: all users working on the same project, but in different work packages)<br /> | |||
• Workflow management<br /> | |||
• Order management<br /> | |||
• Chemical structure handling<br /> | |||
• Dedicated tree structure for storing experiments<br /> | |||
• Legally-binding procedures (signatures)<br /> | |||
• Modular expandability<br /> | |||
• Appropriate integrated analytical features<br /> | |||
• Social networking and collaborative (chat) features<br /> | |||
• Storage for large sets of “raw” data for re-analysis<br /> | |||
|- | |||
|} | |||
|} | |||
==References== | ==References== | ||
Revision as of 17:28, 12 September 2017
| Full article title | Electronic laboratory notebooks in a public–private partnership |
|---|---|
| Journal | PeerJ Computer Science |
| Author(s) | Vaas, Lea A.I.; Witt, Gesa; Windshügel, Björn; Bosin, Andrea; Serra, Giovanni; Bruengger, Adrian; Winterhalter, Mathias; Gribbon, Philip; Levy-Petelinkar, Cindy J.; Kohler, Manfred |
| Author affiliation(s) | Fraunhofer Institute for Molecular Biology and Applied Ecology, University of Cagliari, Basilea Pharmaceutica International AG, Jacobs University Bremen, GlaxoSmithKline |
| Primary contact | Email: manfred dot kohler at ime dot fraunhofer dot de |
| Editors | Baker, Mary |
| Year published | 2016 |
| Volume and issue | 2 |
| Page(s) | e83 |
| DOI | 10.7717/peerj-cs.83 |
| ISSN | 2167-8359 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://peerj.com/articles/cs-83/ |
| Download | https://peerj.com/articles/cs-83.pdf (PDF) |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
This report shares the experience during selection, implementation and maintenance phases of an electronic laboratory notebook (ELN) in a public–private partnership project and comments on users' feedback. In particular, we address which time constraints for roll-out of an ELN exist in granted projects and which benefits and/or restrictions come with out-of-the-box solutions. We discuss several options for the implementation of support functions and potential advantages of open-access solutions. Connected to that, we identified willingness and a vivid culture of data sharing as the major item leading to success or failure of collaborative research activities. The feedback from users turned out to be the only angle for driving technical improvements, but also exhibited high efficiency. Based on these experiences, we describe best practices for future projects on implementation and support of an ELN supporting a diverse, multidisciplinary user group based in academia, NGOs, and/or for-profit corporations located in multiple time zones.
Keywords: public–private partnership, open access, Innovative Medicines Initiative, electronic laboratory notebook, New Drugs for Bad Bugs, IMI, PPP, ND4BB, collaboration, sharing information
Introduction
Laboratory notebooks (LNs) are vital documents of laboratory work in all fields of experimental research. The LN is used to document experimental plans, procedures, results and considerations based on these outcomes. The proper documentation establishes the precedence of results, particularly for inventions of intellectual property (IP). The LN provides the main evidence in the event of disputes relating to scientific publications or patent application. A well-established routine for documentation discourages data falsification by ensuring the integrity of the entries in terms of time, authorship, and content.[1] LNs must be complete, clear, unambiguous and secure. A remarkable example is Alexander Fleming’s documentation, leading to the discovery of penicillin.[2]
The recent development of many novel technologies brought up new platforms in life sciences requiring specialized knowledge. As an example, next-generation sequencing and protein structure determination are generating datasets, which are becoming increasingly prevalent especially in molecular life sciences.[3] The combination and interpretation of these data requires experts from different research areas[4], leading to large research consortia.
In consortia involving multidisciplinary research, the classical paper-based version of a LN is an impediment to efficient data sharing and information exchange. Most of the data from these large-scale collaborative research efforts will never exist in a hard copy format but will be generated in a digitized version. An analysis of this data can be performed by specialized software and dedicated hardware. The classical application of a LN fails in these environments. It is commonly replaced by digital reporting procedures, which can be standardized.[5][6][7] Besides the advantages for daily operational activities, an electronic laboratory notebook (ELN) yields long-term benefits regarding data maintenance. These include, but are not limited to, items listed in Table 1.[8] The order of mentioned points is not expressing any ranking. Besides general tasks, some specific tasks have to be facilitated, especially in the field of drug discovery. One such specific task is searching for chemical structures and substructures in a virtual library of chemical structures and compounds (see Table 1, last item in column “Potentially”). Enabling such a function in an ELN hosting reports about wet-lab work dealing with known drugs and/or compounds to be evaluated would allow dedicated information retrieval for the chemical compounds or (sub-) structures of interest.
| ||||||
Interestingly, although essential for the success of research activities in collaborative settings, the above mentioned advantages are rarely realized by users during daily documentation activities and institutional awareness in academic environment is often lacking.
Since funding agencies and stakeholders are becoming aware of the importance of transparency and reproducibility in both experimental and computational research[9][10][11], the use of digitalized documentation, reproducible analyses and archiving will be a common requirement for funding applications on national and international levels.[12][13][14]
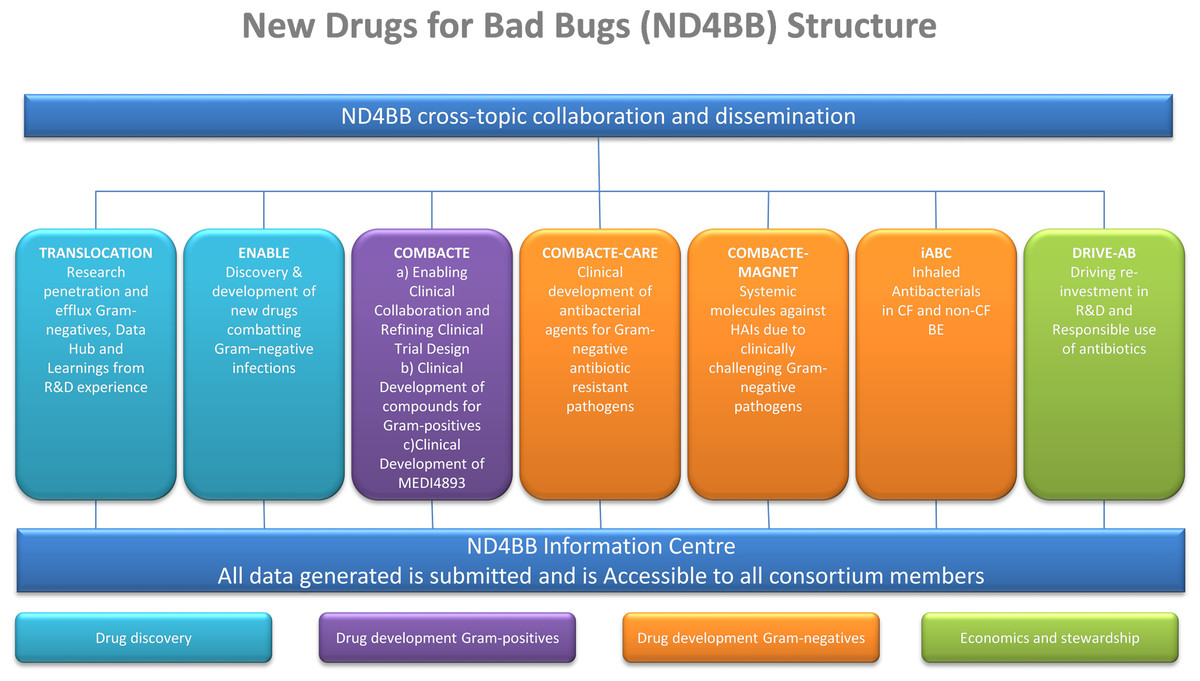
A typical example for a large private-public partnership is the Innovative Medicines Initiative (IMI) New Drugs for Bad Bugs (ND4BB) program[15][16] (see Fig. 1 for details). The program’s objective is to combat antibiotic resistance in Europe by tackling the scientific, regulatory, and business challenges that are hampering the development of new antibiotics.
|
The TRANSLOCATION consortium focus on (i) improving the understanding of the overall permeability of Gram-negative bacteria, and (ii) enhancing the efficiency of antibiotic research and development through knowledge sharing, data sharing and integrated analysis. To meet such complex needs, the TRANSLOCATION consortium was established as a multinational and multisite public–private partnership (PPP) with 15 academic partners, five pharmaceutical companies and seven small- and medium-sized enterprises (SMEs).[17][18][19]
In this article we describe the process of selecting and implementing an ELN in the context of the multisite PPP project TRANSLOCATION, comprising about 90 bench scientists in total. Furthermore we present the results from a survey evaluating the users’ experiences and the benefit for the project two years post-implementation. Based on our experiences, the specific needs in a PPP setting are summarized and lessons learned will be reviewed. As a result, we propose recommendations to assist future users avoiding pitfalls when selecting and implementing ELN software.
Methods
Selection and implementation of an ELN solution
The IMI project call requested a high level of transparency enabling the sharing of data to serve as an example for future projects. The selected consortium TRANSLOCATION had a special demand for an ELN due to its structure — various labs and partners spread widely across Europe needed to report into one common repository — and due to the final goal, data was required to be stored and integrated into one central information hub, the ND4BB Information Centre. Fortunately, no legacy data had to be migrated into the ELN.
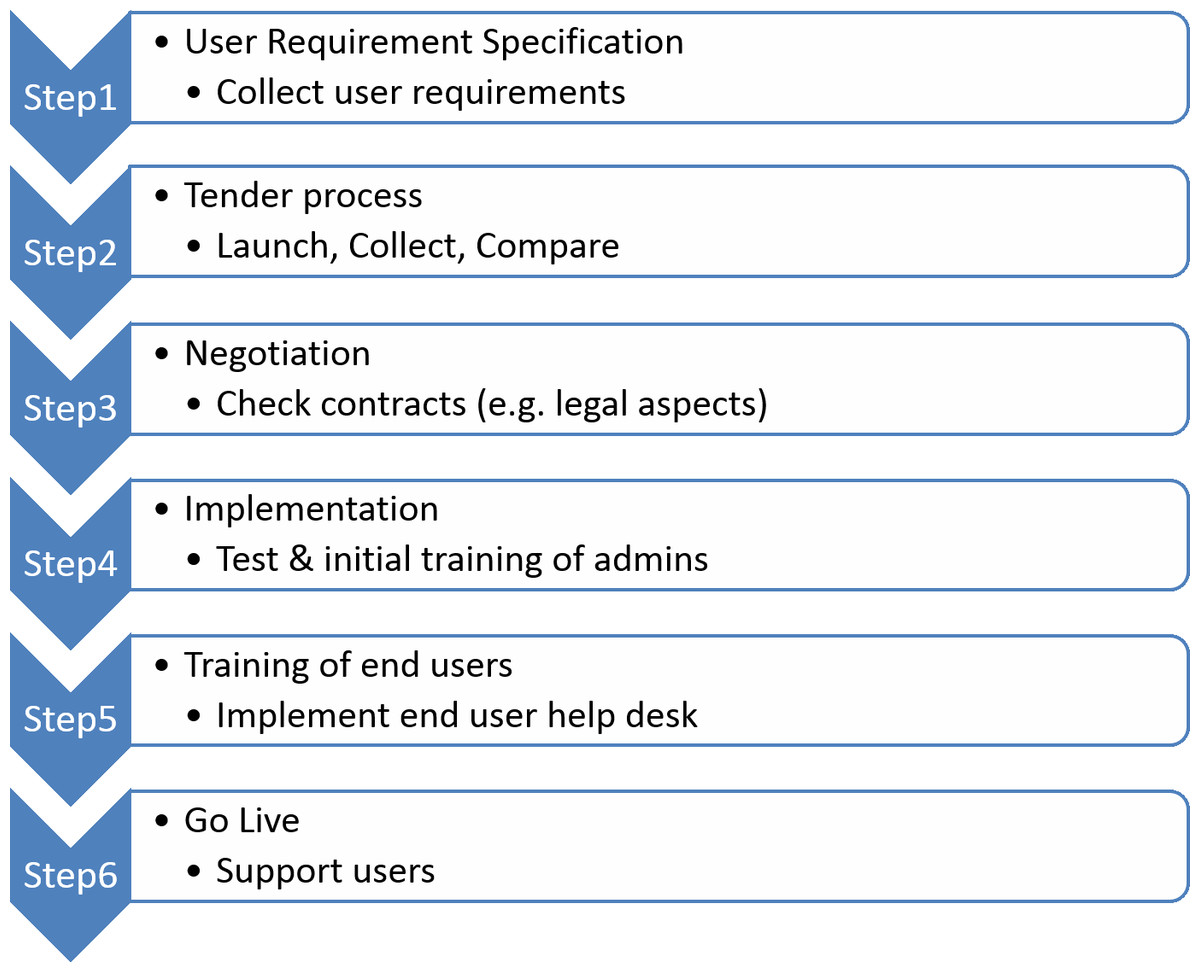
The standard process for the introduction of new software follows a highly structured multi-phase procedure[20][21], as outlined in Fig. 2.
|
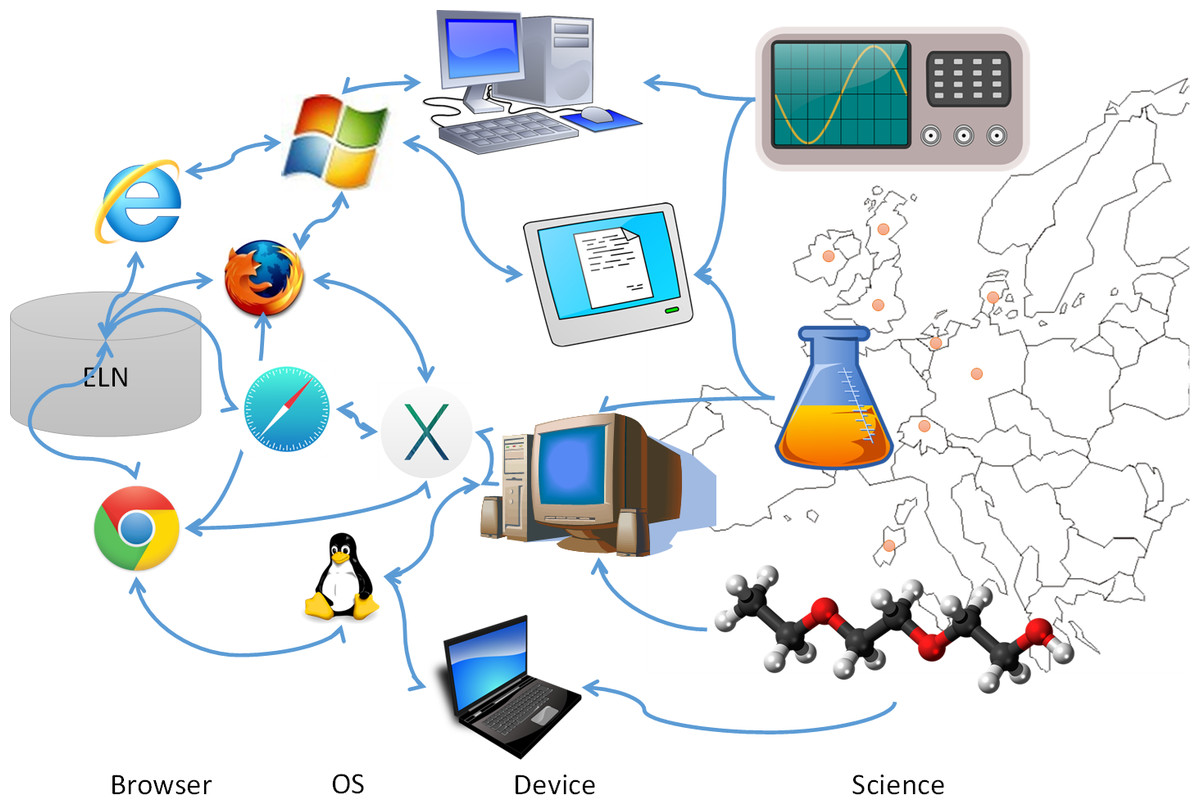
For the first step, we had to manage a large and highly heterogeneous user group (Fig. 3) that would be using the ELN, scheduled for roll-out within six months after project launch. All personnel of the academic partners were requested to enter data into the same ELN, potentially leading to unmet individual user requirements, especially for novices and inexperienced users.
|
As a compromise for step 1 (Fig. 2), we assembled a collection of user requirement specifications (URS) based on the experiences of one laboratory that had already implemented an ELN. We further selected a small group of super users based on their expertise in documentation processes, representing different wet laboratories and in silico environments. The resulting URS was reviewed by IT and business experts from academic as well as private organisations of the consortium. The final version of the URS is available as Supplemental File 1.
In parallel, based on literature (Rubacha, Rattan & Hossel, 2011) and internet searches, presentations of widely used ELNs were evaluated to gain insight into state-of-the-art ELNs. This revealed a wide variety of functional and graphical user interface (GUI) implementations differing in complexity and costs. The continuum between simple out-of-the-box solutions and highly sophisticated and configurable ELNs with interfaces to state-of-the-art analytical tools were covered by the presentations. Notably, the requirements specified by super users also ranged from “easy to use” to “highly individually configurable.” Based on this information, it was clear that the ELN selected for this consortium would never ideally fit all user expectations. Furthermore, the exact number of users and configuration of user groups were unknown at the onset of the project. The most frequently or highest prioritized items of the collected user requirements are listed in Table 2. We divided the gathered requirements into ‘core’ meaning essential and ‘non-core’ meaning ‘nice to have, but not indispensable.’ Further, we list here only the items, which were mentioned by more than two super users from different groups. The full list of URS is available in Supplemental File 1.
| ||||||
References
- ↑ Myers, J.D. (10 July 2014). "Collaborative Electronic Notebooks as Electronic Records: Design Issues for the Secure Electronic Laboratory Notebook (ELN)". ResearchGate. ResearchGate GmbH. https://www.researchgate.net/publication/228705896_Collaborative_electronic_notebooks_as_electronic_records_Design_issues_for_the_secure_electronic_laboratory_notebook_eln. Retrieved 08 January 2015.
- ↑ Bennett, J.W.; Chung, K.T. (2001). "Alexander Fleming and the discovery of penicillin". Advances in Applied Microbiology 49: 163–84. doi:10.1016/S0065-2164(01)49013-7. PMID 11757350.
- ↑ Du, P.; Kofman, J.A. (2007). "Electronic Laboratory Notebooks in Pharmaceutical R&D: On the Road to Maturity". JALA 12 (3): 157–65. doi:10.1016/j.jala.2007.01.001.
- ↑ Ioannidis, J.P.; Greenland, S.; Hlatky, M.A. et al. (2014). "Increasing value and reducing waste in research design, conduct, and analysis". Lancet 383 (9912): 166–75. doi:10.1016/S0140-6736(13)62227-8. PMC PMC4697939. PMID 24411645. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4697939.
- ↑ Special Programme for Research and Training in Tropical Diseases (2006). "Handbook: Quality practices in basic biomedical research". World Health Organization. pp. 122. http://www.who.int/tdr/publications/training-guideline-publications/handbook-quality-practices-biomedical-research/en/.
- ↑ Schnell, S. (2015). "Ten Simple Rules for a Computational Biologist's Laboratory Notebook". PLoS Computational Biology 11 (9): e1004385. doi:10.1371/journal.pcbi.1004385. PMC PMC4565690. PMID 26356732. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4565690.
- ↑ Nussbeck, S.Y.; Weil, P.; Menzel, J. et al. (2014). "The laboratory notebook in the 21st century: The electronic laboratory notebook would enhance good scientific practice and increase research productivity". EMBO Reports 15 (6): 631–4. doi:10.15252/embr.201338358. PMC PMC4197872. PMID 24833749. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4197872.
- ↑ Sandve, G.K.; Nekrutenko, A.; Taylor, J.; Hovig, E. (2013). "Ten simple rules for reproducible computational research". PLoS Computational Biology 9 (10): e1003285. doi:10.1371/journal.pcbi.1003285. PMC PMC3812051. PMID 24204232. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3812051.
- ↑ Bechhofer, S.; Buchan, I.; De Roure, D. et al. (2013). "Why linked data is not enough for scientists". Future Generation Computer Systems 29 (2): 599-611. doi:10.1016/j.future.2011.08.004.
- ↑ White, T.E.; Dalrymple, R.L.; Noble, D.W.A. et al. (2015). "Reproducible research in the study of biological coloration". Animal Behaviour 106: 51–57. doi:10.1016/j.anbehav.2015.05.007.
- ↑ Woelfle, M.; Olliaro, P.; Todd, M.H. (2011). "Open science is a research accelerator". Nature Chemistry 3 (10): 745-8. doi:10.1038/nchem.1149. PMID 21941234.
- ↑ Deutsche Forschungsgemeinschaft (September 2013). "Proposals for Safeguarding Good Scientific Practice" (PDF). https://www.mpimet.mpg.de/fileadmin/publikationen/Volltexte_diverse/DFG-Safeguarding_Good_Scientific_Practice_DFG.pdf.
- ↑ Directorate-General for Research & Innovation (2013). "Guidelines to the Rules on Open Access to Scientific Publications and Open Access to Research Data in Horizon 2020" (PDF). https://ec.europa.eu/research/participants/data/ref/h2020/grants_manual/hi/oa_pilot/h2020-hi-oa-pilot-guide_en.pdf. Retrieved 12 September 2016.
- ↑ Payne, D.J.; Miller, L.F.; Findlay, D. et al. (2015). "Time for a change: addressing R&D and commercialization challenges for antibacterials". Philosophical Transactions of the Royal Society B: Biological Sciences 370 (1670): 20140086. doi:10.1098/rstb.2014.0086.
- ↑ Kostyanev, T.; Bonten, M.J.; O'Brien, S. et al. (2016). "The Innovative Medicines Initiative's New Drugs for Bad Bugs programme: European public-private partnerships for the development of new strategies to tackle antibiotic resistance". Journal of Antimicrobial Chemotherapy 71 (2): 290-5. doi:10.1093/jac/dkv339. PMID 26568581.
- ↑ Rex, J.H. (2014). "ND4BB: addressing the antimicrobial resistance crisis". Nature Reviews Microbiology 12: 231–32. doi:10.1038/nrmicro3245.
- ↑ Stavenger, R.A.; Winterhalter, M. (2014). "TRANSLOCATION project: How to get good drugs into bad bugs". Science Translational Medicine 6 (228): 228ed7. doi:10.1126/scitranslmed.3008605. PMID 24648337.
- ↑ "ND4BB Translocation". ND4BB. http://translocation.eu/. Retrieved 22 September 2015.
- ↑ 20.0 20.1 Nehme, A.; Scoffin, R.A. (2006). "9. Electronic Laboratory Notebooks". In Ekins, S.; Wang, B.. Computer Applications in Pharmaceutical Research and Development. Wiley. pp. 209–27. ISBN 9780471737797.
- ↑ Milsted, A.J.; Hale, J.R.; Grey, J.G.; Neylon, C. (2013). "LabTrove: a lightweight, web based, laboratory "blog" as a route towards a marked up record of work in a bioscience research laboratory". PLoS One 8 (7): e67460. doi:10.1371/journal.pone.0067460. PMC PMC3720848. PMID 23935832. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3720848.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation, including regionalizing spelling. In some cases important information was missing from the references, and that information was added. The original lists references in alphabetical order; this version lists them in order of appearance due to the nature of the wiki.