Difference between revisions of "Journal:Evidence-based design and evaluation of a whole genome sequencing clinical report for the reference microbiology laboratory"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) (Saving and adding more.) |
||
| Line 47: | Line 47: | ||
Work in the [[electronic health record]] (EHR) and patient risk communication domains has also provided insight into not just the final product but also the process of effective design. Through quantitative and qualitative evaluations, research has shown that some EHRs are difficult to use because they were not designed to support clinical tasks and information retrieval, but rather data entry.<ref name="WrightHow98" /> Reviews of the risk communication literature note that while many visual aids improve patients’ understanding of risk<ref name="ZipkinEvidence14">{{cite journal |title=Evidence-based risk communication: A systematic review |journal=Annals of Internal Medicine |author=Zipkin, D.A.; Umscheid, C.A.; Keating, N.L. et al. |volume=161 |issue=4 |pages=270-80 |doi=10.7326/M14-0295 |pmid=25133362}}</ref>, the design features that viewers preferred — namely simplistic, minimalist designs — were not necessarily those that led to an accurate interpretation of the underlying data.<ref name="AnckerDesign06">{{cite journal |title=Design features of graphs in health risk communication: A systematic review |journal=JAMIA |author=Ancker, J.S.; Senathirajah, Y.; Kukafka, R. et al. |volume=13 |issue=6 |pages=608–18 |doi=10.1197/jamia.M2115 |pmid=16929039 |pmc=PMC1656964}}</ref> Together, these gaps indicate a need for a human-centered, participatory approach iteratively incorporating both design and evaluation.<ref name="HettingerCog17">{{cite journal |title=Cognitive engineering and health informatics: Applications and intersections |journal=Journal of Biomedical Informatics |author=Hettinger, A.Z.; Roth, E.M.; Bisantz, A.M. |volume=67 |pages=21–33 |doi=10.1016/j.jbi.2017.01.010 |pmid=28126605}}</ref><ref name="HorskyInterface12">{{cite journal |title=Interface design principles for usable decision support: A targeted review of best practices for clinical prescribing interventions |journal=Journal of Biomedical Informatics |author=Horsky, J.; Schiff, G.D.; Johnston, D. et al. |volume=45 |issue=6 |pages=1202-16 |doi=10.1016/j.jbi.2012.09.002 |pmid=22995208}}</ref> | Work in the [[electronic health record]] (EHR) and patient risk communication domains has also provided insight into not just the final product but also the process of effective design. Through quantitative and qualitative evaluations, research has shown that some EHRs are difficult to use because they were not designed to support clinical tasks and information retrieval, but rather data entry.<ref name="WrightHow98" /> Reviews of the risk communication literature note that while many visual aids improve patients’ understanding of risk<ref name="ZipkinEvidence14">{{cite journal |title=Evidence-based risk communication: A systematic review |journal=Annals of Internal Medicine |author=Zipkin, D.A.; Umscheid, C.A.; Keating, N.L. et al. |volume=161 |issue=4 |pages=270-80 |doi=10.7326/M14-0295 |pmid=25133362}}</ref>, the design features that viewers preferred — namely simplistic, minimalist designs — were not necessarily those that led to an accurate interpretation of the underlying data.<ref name="AnckerDesign06">{{cite journal |title=Design features of graphs in health risk communication: A systematic review |journal=JAMIA |author=Ancker, J.S.; Senathirajah, Y.; Kukafka, R. et al. |volume=13 |issue=6 |pages=608–18 |doi=10.1197/jamia.M2115 |pmid=16929039 |pmc=PMC1656964}}</ref> Together, these gaps indicate a need for a human-centered, participatory approach iteratively incorporating both design and evaluation.<ref name="HettingerCog17">{{cite journal |title=Cognitive engineering and health informatics: Applications and intersections |journal=Journal of Biomedical Informatics |author=Hettinger, A.Z.; Roth, E.M.; Bisantz, A.M. |volume=67 |pages=21–33 |doi=10.1016/j.jbi.2017.01.010 |pmid=28126605}}</ref><ref name="HorskyInterface12">{{cite journal |title=Interface design principles for usable decision support: A targeted review of best practices for clinical prescribing interventions |journal=Journal of Biomedical Informatics |author=Horsky, J.; Schiff, G.D.; Johnston, D. et al. |volume=45 |issue=6 |pages=1202-16 |doi=10.1016/j.jbi.2012.09.002 |pmid=22995208}}</ref> | ||
===Collaboration context—COMPASS-TB=== | |||
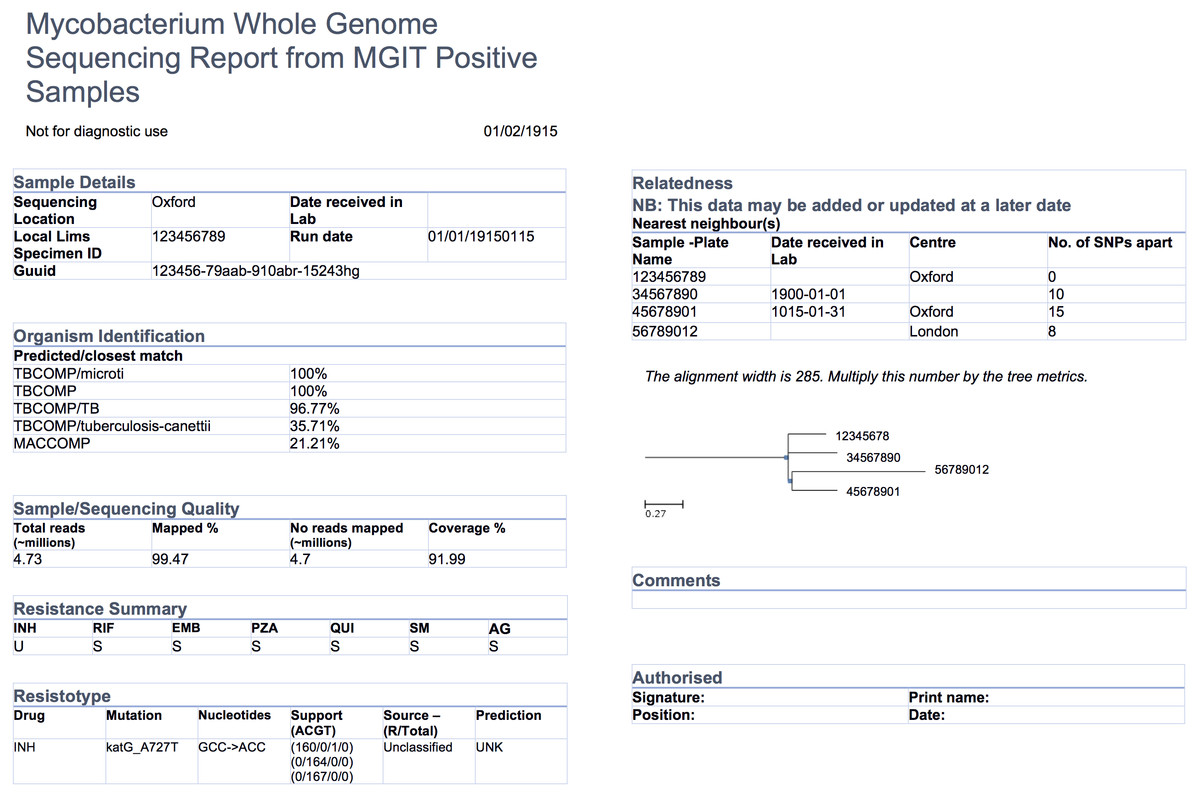
The COMPASS-TB project was a proof-of-concept study demonstrating the feasibility and utility of WGS for diagnosing tuberculosis (TB) infection, evaluating an isolate’s antimicrobial sensitivity/resistance and genotyping the isolate to identify epidemiologically related cases.<ref name="PankhurstRapid16" /> On the basis of COMPASS-TB’s results, Public Health England (PHE) has implemented routine WGS in the TB reference laboratory<ref name="PHETuber16">{{cite web |url=https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/654294/TB_Annual_Report_2016_GTW2309_errata_v1.2.pdf |format=PDF |title=Tuberculosis in England: 2016 Report |publisher=Public Health England |date=September 2016}}</ref>; however, this requires changing how mycobacteriology results are reported to clinical and public health stakeholders. The COMPASS-TB pilot used reports designed by the project team, but as clinical implementation within PHE progressed, team members expressed an interest in redesigning the report (Fig. 1) to facilitate interpretation of this new data type and align [[laboratory]] reporting practices with the needs of multiple TB stakeholders. | |||
[[File:Fig1 Crisan PeerJ2018 6.jpg|1000px]] | |||
{{clear}} | |||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| border="0" cellpadding="5" cellspacing="0" width="1000px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Fig. 1''' An earlier COMPASS-TB report design</blockquote> | |||
|- | |||
|} | |||
|} | |||
==References== | ==References== | ||
Revision as of 17:21, 15 February 2018
| Full article title | Evidence-based design and evaluation of a whole genome sequencing clinical report for the reference microbiology laboratory |
|---|---|
| Journal | PeerJ |
| Author(s) | Crisan, Anamaria; McKee, Geoffrey; Munzner, Tamara; Gardy, Jennifer L. |
| Author affiliation(s) | University of British Columbia, British Columbia Centre for Disease Control |
| Primary contact | Email: jennifer dot gardy at bccdc dot ca |
| Editors | Smidt, H. |
| Year published | 2018 |
| Volume and issue | 6 |
| Page(s) | e4218 |
| DOI | 10.7717/peerj.4218 |
| ISSN | 2167-8359 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://peerj.com/articles/4218/ |
| Download | https://peerj.com/articles/4218.pdf (PDF) |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
Background: Microbial genome sequencing is now being routinely used in many clinical and public health laboratories. Understanding how to report complex genomic test results to stakeholders who may have varying familiarity with genomics — including clinicians, laboratorians, epidemiologists, and researchers — is critical to the successful and sustainable implementation of this new technology; however, there are no evidence-based guidelines for designing such a report in the pathogen genomics domain. Here, we describe an iterative, human-centered approach to creating a report template for communicating tuberculosis (TB) genomic test results.
Methods: We used design study methodology — a human centered approach drawn from the information visualization domain — to redesign an existing clinical report. We used expert consults and an online questionnaire to discover various stakeholders’ needs around the types of data and tasks related to TB that they encounter in their daily workflow. We also evaluated their perceptions of and familiarity with genomic data, as well as its utility at various clinical decision points. These data shaped the design of multiple prototype reports that were compared against the existing report through a second online survey, with the resulting qualitative and quantitative data informing the final, redesigned, report.
Results: We recruited 78 participants, 65 of whom were clinicians, nurses, laboratorians, researchers, and epidemiologists involved in TB diagnosis, treatment, and/or surveillance. Our first survey indicated that participants were largely enthusiastic about genomic data, with the majority agreeing on its utility for certain TB diagnosis and treatment tasks and many reporting some confidence in their ability to interpret this type of data (between 58.8% and 94.1%, depending on the specific data type). When we compared our four prototype reports against the existing design, we found that for the majority (86.7%) of design comparisons, participants preferred the alternative prototype designs over the existing version, and that both clinicians and non-clinicians expressed similar design preferences. Participants showed clearer design preferences when asked to compare individual design elements versus entire reports. Both the quantitative and qualitative data informed the design of a revised report, available online as a LaTeX template.
Conclusions: We show how a human-centered design approach integrating quantitative and qualitative feedback can be used to design an alternative report for representing complex microbial genomic data. We suggest experimental and design guidelines to inform future design studies in the bioinformatics and microbial genomics domains. We also suggest that this type of mixed-methods study is important to facilitate the successful translation of pathogen genomics in the clinic, not only for clinical reports but also more complex bioinformatics data visualization software.
Keywords: human-centered design, next-generation sequencing, report, tuberculosis, genome
Introduction
Whole genome sequencing (WGS) is quickly moving from proof-of-concept research into routine clinical and public health use. WGS can diagnose infections at least as accurately as current protocols[1][2], can predict antimicrobial resistance phenotypes for certain drugs[3][4][5] with high concordance to culture-based testing methods, and can be used in outbreak surveillance to resolve transmission clusters at a resolution not possible with existing genomic or epidemiological methods.[6] Importantly, WGS offers faster turnaround times compared to many culture-based tests, particularly for antimicrobial resistance testing in slow-growing bacteria.
As reference microbiology laboratories move towards accreditation of WGS for routine clinical use, the community is turning its attention toward standardization, developing standard operating procedures for reproducible sample handling, sequencing, and downstream bioinformatics analysis.[7][8] Reporting genomic microbiology test results in a way that is interpretable by clinicians, nurses, laboratory staff, researchers, and surveillance experts and that meets regulatory requirements is equally important; however, relatively little effort has been directed toward this area. WGS clinical reports are often produced in-house on an ad hoc, project-by-project basis, with the resulting product not necessarily meeting the needs of the many stakeholders using the report in their clinical and surveillance workflows.
Human-centered design in the clinical laboratory
The information visualization, human–computer interaction, and usability engineering fields offer techniques and design guidelines that have informed bioinformatics tools, including Disease View[9] for exploring host-pathogen interaction data and Microreact[10] for visualizing phylogenetic trees in the context of epidemiological or clinical data. Although the public health community is beginning to recognize the potential role of visualization and analytics in daily laboratory workflows[11] these techniques have not yet been applied to routine reporting of microbiological test results. However, work from the human health domain — particularly the formatting and display of pathology reports, where standardization is critical[12] — sheds light on the complex task of clinical report design.
Valenstein reports four principles for organizing an effective pathology report: use headlines to emphasize key points, ensure design continuity over time and relative to other reports, consider information density, and reduce clutter[13], while Renshaw et al.[14] note that when pathology report templates were reformatted with numbering and bolding to highlight required information, template completion rates rose from 84 to 98%. Fixed, consistent layout of medical record elements, highlighting of data relative to background text, and single-page layout improve clinicians’ ability to locate information[15], while information design principles, including visually structuring the document to separate different elements and organizing information to meet the needs of multiple stakeholder types, can reduce the number of errors in data interpretation.[16]
Work in the electronic health record (EHR) and patient risk communication domains has also provided insight into not just the final product but also the process of effective design. Through quantitative and qualitative evaluations, research has shown that some EHRs are difficult to use because they were not designed to support clinical tasks and information retrieval, but rather data entry.[16] Reviews of the risk communication literature note that while many visual aids improve patients’ understanding of risk[17], the design features that viewers preferred — namely simplistic, minimalist designs — were not necessarily those that led to an accurate interpretation of the underlying data.[18] Together, these gaps indicate a need for a human-centered, participatory approach iteratively incorporating both design and evaluation.[19][20]
Collaboration context—COMPASS-TB
The COMPASS-TB project was a proof-of-concept study demonstrating the feasibility and utility of WGS for diagnosing tuberculosis (TB) infection, evaluating an isolate’s antimicrobial sensitivity/resistance and genotyping the isolate to identify epidemiologically related cases.[4] On the basis of COMPASS-TB’s results, Public Health England (PHE) has implemented routine WGS in the TB reference laboratory[21]; however, this requires changing how mycobacteriology results are reported to clinical and public health stakeholders. The COMPASS-TB pilot used reports designed by the project team, but as clinical implementation within PHE progressed, team members expressed an interest in redesigning the report (Fig. 1) to facilitate interpretation of this new data type and align laboratory reporting practices with the needs of multiple TB stakeholders.
|
References
- ↑ Fukui, Y.; Aoki, K.; Okuma, S. et al.. "Metagenomic analysis for detecting pathogens in culture-negative infective endocarditis". Journal of Infection and Chemotherapy 21 (12): 882–4. doi:10.1016/j.jiac.2015.08.007. PMID 26360016.
- ↑ Loman, N.J.; Constantinidou, C.; Christner, M. et al.. "A culture-independent sequence-based metagenomics approach to the investigation of an outbreak of Shiga-toxigenic Escherichia coli O104:H4". JAMA 309 (14): 1502-10. doi:10.1001/jama.2013.3231. PMID 23571589.
- ↑ Bradley, P.; Gordon, N.C.; Walker, T.M. et al.. "Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis". Nature Communications 6: 10063. doi:10.1038/ncomms10063. PMC PMC4703848. PMID 26686880. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4703848.
- ↑ 4.0 4.1 Pankhurst, L.J.; Del Ojo Elias, C.; Votintseva, A.A. et al.. "Rapid, comprehensive, and affordable mycobacterial diagnosis with whole-genome sequencing: A prospective study". The Lancet Respiratory Medicine 4 (1): 49-58. doi:10.1016/S2213-2600(15)00466-X. PMC PMC4698465. PMID 26669893. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4698465.
- ↑ Walker, T.M.; Kohl, T.A.; Omar, S.V. et al.. "Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: A retrospective cohort study". The Lancet Infectious Diseases 15 (10): 1193–1202. doi:10.1016/S1473-3099(15)00062-6. PMC PMC4579482. PMID 26116186. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4579482.
- ↑ Nikolayevskyy, V.; Kranzer, K.; Niemann, S. et al.. "Whole genome sequencing of Mycobacterium tuberculosis for detection of recent transmission and tracing outbreaks: A systematic review". Tuberculosis 98: 77-85. doi:10.1016/j.tube.2016.02.009. PMID 27156621.
- ↑ Budowle, B.; Connell, N.D.; Bielecka-Oder, A. et al.. "Validation of high throughput sequencing and microbial forensics applications". Investigative Genetics 5: 9. doi:10.1186/2041-2223-5-9. PMC PMC4123828. PMID 25101166. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4123828.
- ↑ Gargis, A.S.; Kalman, L.; Lubin, I.M.. "Assuring the Quality of Next-Generation Sequencing in Clinical Microbiology and Public Health Laboratories". Journal of Clinical Microbiology 54 (12): 2857-2865. doi:10.1128/JCM.00949-16. PMC PMC5121372. PMID 27510831. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5121372.
- ↑ Driscoll, T.; Gabbard, J.L.; Mao, C. et al.. "Integration and visualization of host-pathogen data related to infectious diseases". Bioinformatics 27 (16): 2279-87. doi:10.1093/bioinformatics/btr391. PMC PMC3150046. PMID 21712250. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3150046.
- ↑ Argimón, S.; Abudahab, K.; Goater, R.J. et al.. "Microreact: Visualizing and sharing data for genomic epidemiology and phylogeography". Microbial Genomics 2 (11): e000093. doi:10.1099/mgen.0.000093. PMC PMC5320705. PMID 28348833. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5320705.
- ↑ Carroll, L.N.; Au, A.P.; Detwiler, L.T. et al.. "Visualization and analytics tools for infectious disease epidemiology: A systematic review". Journal of Biomedical Informatics 51: 287-98. doi:10.1016/j.jbi.2014.04.006. PMC PMC5734643. PMID 24747356. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5734643.
- ↑ Leslie, K.O.; Rosai, J.. "Standardization of the surgical pathology report: Formats, templates, and synoptic reports". Seminars in Diagnostic Pathology 11 (4): 253-7. PMID 7878300.
- ↑ Valenstein, P.N.. "Formatting pathology reports: Applying four design principles to improve communication and patient safety". Archives of Pathology and Laboratory Medicine 132 (1): 84-94. doi:0.1043/1543-2165(2008)132[84:FPRAFD]2.0.CO;2. PMID 18181680.
- ↑ Renshaw, S.A.; Mena-Allauca, M.; Touriz, M. et al.. "The impact of template format on the completeness of surgical pathology reports". Archives of Pathology and Laboratory Medicine 138 (1): 121-4. doi:10.5858/arpa.2012-0733-OA. PMID 24377820.
- ↑ Nygren, E.; Wyatt, J.C.; Wright, P.. "Helping clinicians to find data and avoid delays". Lancet 352 (9138): 1462-6. doi:10.1016/S0140-6736(97)08307-4. PMID 9808009.
- ↑ 16.0 16.1 Wright, P.; Jansen, C.; Wyatt, J.C.. "How to limit clinical errors in interpretation of data". Lancet 352 (9139): 1539-43. doi:10.1016/S0140-6736(98)08308-1. PMID 9820319.
- ↑ Zipkin, D.A.; Umscheid, C.A.; Keating, N.L. et al.. "Evidence-based risk communication: A systematic review". Annals of Internal Medicine 161 (4): 270-80. doi:10.7326/M14-0295. PMID 25133362.
- ↑ Ancker, J.S.; Senathirajah, Y.; Kukafka, R. et al.. "Design features of graphs in health risk communication: A systematic review". JAMIA 13 (6): 608–18. doi:10.1197/jamia.M2115. PMC PMC1656964. PMID 16929039. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1656964.
- ↑ Hettinger, A.Z.; Roth, E.M.; Bisantz, A.M.. "Cognitive engineering and health informatics: Applications and intersections". Journal of Biomedical Informatics 67: 21–33. doi:10.1016/j.jbi.2017.01.010. PMID 28126605.
- ↑ Horsky, J.; Schiff, G.D.; Johnston, D. et al.. "Interface design principles for usable decision support: A targeted review of best practices for clinical prescribing interventions". Journal of Biomedical Informatics 45 (6): 1202-16. doi:10.1016/j.jbi.2012.09.002. PMID 22995208.
- ↑ "Tuberculosis in England: 2016 Report" (PDF). Public Health England. September 2016. https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/654294/TB_Annual_Report_2016_GTW2309_errata_v1.2.pdf.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In several cases the PubMed ID was missing and was added to make the reference more useful. The original article lists references alphabetically, but this version — by design — lists them in order of appearance.