Journal:Laboratory testing methods for novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)
| Full article title | Laboratory testing methods for novel Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-CoV-2) |
|---|---|
| Journal | Frontiers in Cell and Developmental Biology |
| Author(s) | D'Cruz, Roshan J.; Currier, Arthur W.; Sampson, Valerie B. |
| Author affiliation(s) | Nemours/Alfred I. duPont Hospital for Children |
| Primary contact | Email: valerie dot sampson at nemours dot org |
| Year published | 2020 |
| Volume and issue | 8 |
| Article # | 468 |
| DOI | 10.3389/fcell.2020.00468 |
| ISSN | 2296-634X |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.frontiersin.org/articles/10.3389/fcell.2020.00468/full |
| Download | https://www.frontiersin.org/articles/10.3389/fcell.2020.00468/pdf (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
Following the first reports of coronavirus disease 2019 (COVID-19) by China to the World Health Organization (WHO) on December 31, 2019, more than 4,302,774 cases of infection by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus have been reported by authorities in 212 countries and territories as of May 12, 2020. The outbreak and spread of COVID-19 worldwide highlights the critical need for developing rapid and accurate diagnostic testing methods for emerging human coronavirus (CoV) infections. Testing is crucial to tracking the spread of disease during a pandemic, and to swiftly permitting public health interventions, including isolation, quarantine, and appropriate clinical management of afflicted individuals. The key components of viral diagnostic tests are (1) collection of the appropriate sample (blood, nasal swab, and throat swab); (2) availability of the genetic and proteomic sequences of the novel virus for analysis; and (3) rapid and accurate laboratory testing methods. The current gold standard for the molecular diagnosis of SARS-CoV-2 infection is the real-time reverse transcription polymerase chain reaction (qRT-PCR) for the qualitative and quantitative detection of viral nucleic acids. Other relevant laboratory methods include enzyme-linked immunosorbent assays (ELISA) for viral antibody and antigen detection, and serum virus neutralization (SVN) assays for antibody neutralization determination. The challenges faced in developing a diagnostic test for a novel pathogen are the ability to measure low viral loads for early detection, to provide low or no cross-reactivity with other viral strains, and to deliver results rapidly. Several point-of-care molecular devices are currently being integrated for fast and accurate diagnosis of SARS-CoV-2 infections. This review discusses the current laboratory methods available to test for coronaviruses by focusing on the present COVID-19 outbreak.
Keywords: coronavirus, RT-PCR, ELISA, lateral flow diagnostics, convalescent plasma
Introduction
Coronavirus disease 2019 (COVID-19) is caused by a novel coronavirus (CoV) that was originally reported in Wuhan, Hubei province, China in December 2019.[1] The International Committee on Taxonomy of Viruses named the virus "severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Infection by SARS-CoV-2 causes a respiratory illness that varies in severity from mild upper respiratory symptoms akin to the seasonal flu, to severe progressive respiratory failure that requires intensive care and can lead to death. Asymptomatic carriers of the virus have also been reported and pose a significant public health threat due to their ability to unknowingly spread the virus.[2] SARS-CoV-2 represents the third coronavirus in this millennium to cross species from animals to humans and cause a severe respiratory disease, after Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012[3] and severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003.[4][5] SARS-CoV-2 has now been identified as the seventh coronavirus that is transmissible between humans (a group which also includes HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1).[6] On January 30, 2020, the World Health Organization (WHO) declared the SARS-CoV-2 epidemic a public health emergency of international concern, and the public health emergency was upgraded to a pandemic on March 11. At least 4,302,774 confirmed cases and 289,561 deaths worldwide were reported as of May 12.[7] Diagnostic testing is critical during a pandemic as the ability to track the spread of SARS-CoV-2 is essential for effective disease management and control.
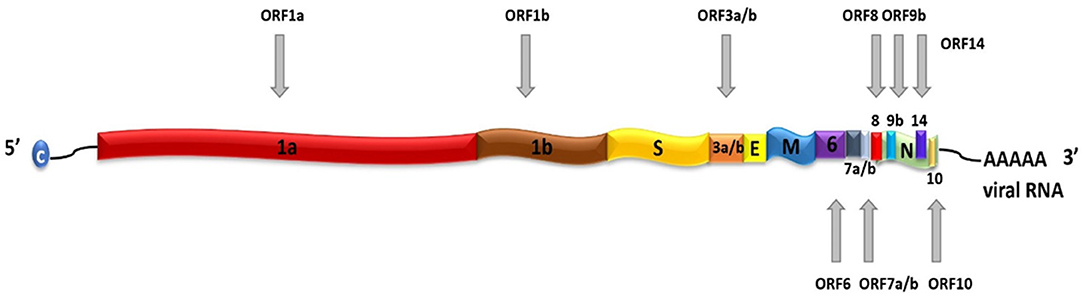
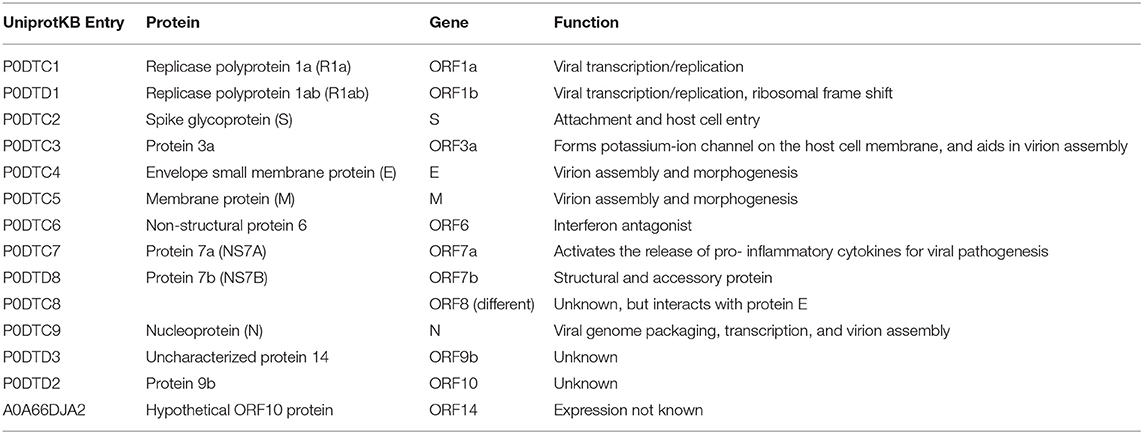
SARS-CoV-2 is a positive-sense, single-stranded RNA (ssRNA), group IV virus. The genome was sequenced from the bronchoalveolar lavage fluid of a patient (Genbank: MN908947) and shared through the Global Initiative on Sharing All Influenza Data (GISAID) platform on January 12, 2020.[8] The ~30 k base pair genome is highly similar to the human SARS-CoV and bat CoV-SARS-like genomes, with 14 open reading frames (ORFs) that encode structural, replication, and non-structural accessory proteins, as depicted in Figure 1. Molecular modeling studies demonstrate that like SARS-CoV, SARS-CoV-2 is surrounded by a lipid bilayer membrane, containing structural membrane (M) and envelope (E) proteins that interact to form the viral envelope.[9] This layer also contains spike glycoproteins (S) that give the characteristic “corona” appearance of this family of viruses. The spike proteins bind specific host cell receptors to facilitate host cell attachment and entry.[10] The nucleic acid-associated protein binds the RNA genome and forms the nucleocapsid (N). Other proteins include replication and non-structural accessory proteins, listed in Table 1. Reports of different strains of SARS-CoV-2 suggest an early split from the SARS-CoV-2 lineage and/or that the virus is mutating. Ongoing research provides insight into the unique and conserved features of the genome and proteome of SARS-CoV-2 for tracking mutations and generates evidence about the evolution of the virus.[11][12] This is important as these changes may affect key structural and non-structural components of SARS-CoV-2 that can render some diagnostic tests ineffective or less sensitive and can also impact the selection of epitopes for the development of new tests.
|
|
References
- ↑ World Health Organization (2020). "Naming the coronavirus disease (COVID-19) and the virus that causes it". World Health Organization. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it. Retrieved May 2020.
- ↑ Chan, J.F.-W.; Yuan, S.; Kok, K.-H. et al. (2020). "A Familial Cluster of Pneumonia Associated With the 2019 Novel Coronavirus Indicating Person-To-Person Transmission: A Study of a Family Cluster". Lancet 395 (10223): 514–23. doi:10.1016/S0140-6736(20)30154-9. PMC PMC7159286. PMID 31986261. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7159286.
- ↑ Zaki, A.M.; van Boheemen, S.; Besterbroer, T.M. et al. (2012). "Isolation of a Novel Coronavirus From a Man With Pneumonia in Saudi Arabia". New England Journal of Medicine 367 (19): 1814–20. doi:10.1056/NEJMoa1211721. PMID 23075143.
- ↑ Drosten, C.; Günther, S.; Preiser, W. et al. (2003). "Identification of a Novel Coronavirus in Patients With Severe Acute Respiratory Syndrome". New England Journal of Medicine 348 (20): 1967-76. doi:10.1056/NEJMoa030747. PMID 12690091.
- ↑ Ksiazek, T.G.; Erdman, D.; Goldsmith, C.S. et al. (2003). "A Novel Coronavirus Associated With Severe Acute Respiratory Syndrome". New England Journal of Medicine 348 (20): 1953–66. doi:10.1056/NEJMoa030781. PMID 12690092.
- ↑ Salata, C.; Calistri, A.; Parolin, C. et al. (2019). "Coronaviruses: A Paradigm of New Emerging Zoonotic Diseases". Pathogens and Disease 77 (9): ftaa006. doi:10.1093/femspd/ftaa006. PMC PMC7108526. PMID 32065221. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7108526.
- ↑ "COVID-19 Coronavirus Pandemic". Worldometer. https://www.worldometers.info/coronavirus/. Retrieved 12 May 2020. >
- ↑ Wu, F.; Zhao, S.; Yu, B. et al. (2020). "A New Coronavirus Associated With Human Respiratory Disease in China". Nature 579 (7798): 265-269. doi:10.1038/s41586-020-2008-3. PMC PMC7094943. PMID 32015508. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7094943.
- ↑ Durrant, J.D.; Kochanek, S.E.; Casalino, L. et al. (2020). "Mesoscale All-Atom Influenza Virus Simulations Suggest New Substrate Binding Mechanism". ACS Central Science 6 (2): 189–96. doi:10.1021/acscentsci.9b01071. PMC PMC7048371. PMID 32123736. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7048371.
- ↑ Graham, R.L.; Baric, R.S. (2010). "Recombination, Reservoirs, and the Modular Spike: Mechanisms of Coronavirus Cross-Species Transmission". Journal of Virology 84 (7): 3134–46. doi:10.1128/JVI.01394-09. PMC PMC2838128. PMID 19906932. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2838128.
- ↑ Phan, T. (2020). "Genetic Diversity and Evolution of SARS-CoV-2". Infection, genetics and evolution 81: 104260. doi:10.1016/j.meegid.2020.104260. PMC PMC7106203. PMID 32092483. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7106203.
- ↑ Wang, C.; Liu, Z.; Chen, Z. et al. (2020). "The Establishment of Reference Sequence for SARS-CoV-2 and Variation Analysis". Journal of Medical Virology 92 (6): 667-674. doi:10.1002/jmv.25762. PMC PMC7228400. PMID 32167180. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7228400.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. References in this version are listed in order of appearance—by design—rather than alphabetical order as the original was.