Difference between revisions of "Journal:OpenBIS ELN-LIMS: An open-source database for academic laboratories"
Shawndouglas (talk | contribs) (Created stub. Saving and adding more.) |
Shawndouglas (talk | contribs) (Saving and adding more.) |
||
| Line 33: | Line 33: | ||
Several commercial database solutions for ELNs are available, but they are not commonly adopted by academia. An internal evaluation among the research groups of the departments of Biosystems Science and Engineering and Biology of ETH Zurich found that the options affordable to academic labs had either unsuitable user interfaces (UI) or lack of features. The combination of an easy-to-use UI, flexible adaptation to a labs needs, support of an [[audit trail]], physical support of the data server, support for managing large amounts of data and for integrating new measurement devices was only available at very high costs. On the other hand, the available open-source platforms lack many of the above features and, as they often originate from a single person within a research group, they also lack maintenance and support during the required data management life cycle. | Several commercial database solutions for ELNs are available, but they are not commonly adopted by academia. An internal evaluation among the research groups of the departments of Biosystems Science and Engineering and Biology of ETH Zurich found that the options affordable to academic labs had either unsuitable user interfaces (UI) or lack of features. The combination of an easy-to-use UI, flexible adaptation to a labs needs, support of an [[audit trail]], physical support of the data server, support for managing large amounts of data and for integrating new measurement devices was only available at very high costs. On the other hand, the available open-source platforms lack many of the above features and, as they often originate from a single person within a research group, they also lack maintenance and support during the required data management life cycle. | ||
Here, we describe the open-source database openBIS (open Biology Information System) electronic laboratory notebook and a laboratory information management system (ELN-LIMS), a software specifically developed with academic life science labs in mind. The structure of the database ensures that the results are represented in a logical and meaningful fashion, and it guarantees that the users can find, understand and reuse the data. openBIS ELN-LIMS consists of two interconnected parts: the LIMS, where the information about materials and methods is stored and kept up to date and the ELN, where data obtained from experiments are uploaded and annotated. The system is based on the openBIS platform<ref name="Bauch_openBIS11">{{cite journal |title=openBIS: A flexible framework for managing and analyzing complex data in biology research |journal=BMC Bioinformatics |author=Bauch, A.; Adamczyk, I.; Buczek, P. et al. |volume=12 |pages=468 |year=2011 |doi=10.1186/1471-2105-12-468 |pmid=22151573 |pmc=PMC3275639}}</ref>, which, since 2007, is actively developed and maintained by a dedicated team of software engineers at the ETH and is productively used in several academic institutions and a few private companies. | |||
==Results== | |||
===System's overview=== | |||
Our goal was to implement a database to support the research activity of a typical biology laboratory. We conceived it as a tool to assist everyday activities: storage of materials, methods set-up, design and description of experiments, data labeling, storage and organization of results. To this purpose, we based our system on the platform openBIS<ref name="Bauch_openBIS11" />, as it has the necessary features described above, is field-tested since about seven years and known to work well also with hundreds of terabytes of data. | |||
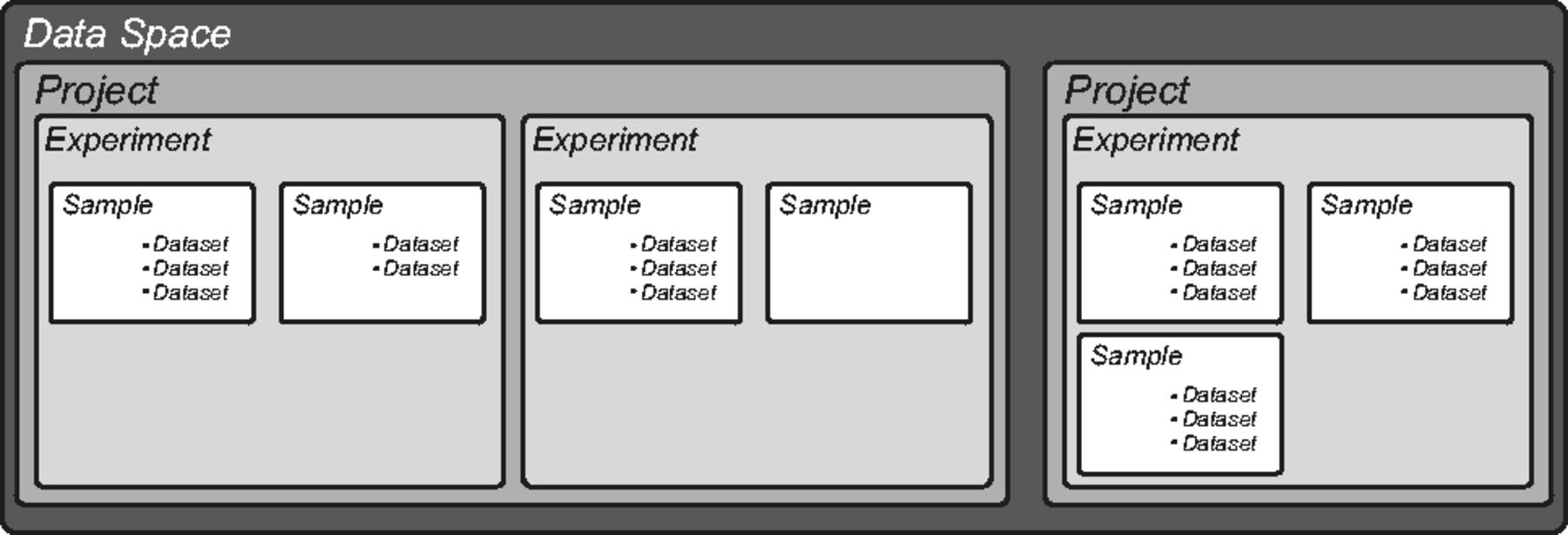
The openBIS core has a predefined hierarchical structure (Fig. 1). The top level, called "Data Space", controls access and rights and contains one or more "Projects," which in turn contain one or more "Experiments." "Samples" are the basic unit of openBIS and are organized within Experiments. We adapted this hierarchical structure to describe the organization and the objects of a typical life science laboratory. We defined the type and the number of Experiments and Samples and assigned customized properties to describe metadata. External files, in any kind of format, are uploaded as "Datasets" to each Sample. Each Sample can have any number of Datasets. We defined different types of Datasets and defined appropriate sets of properties (or annotations). | |||
[[File:Fig1 Barillari Bioinformatics2015 32-4.jpg|800px]] | |||
{{clear}} | |||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| border="0" cellpadding="5" cellspacing="0" width="800px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 1.''' Hierarchical structure of openBIS. The levels are indicated with different grey shades.</blockquote> | |||
|- | |||
|} | |||
|} | |||
We used parent–child relationships linking Samples or Datasets to describe the logical or physical connections existing between the corresponding objects in the real world. We introduced the possibility to describe the nature of these relationships by annotating the links using texts or keywords. | |||
To ensure that the creation, modification or deletion of any electronic records is traceable, computer-generated, time-stamped audit trails are used to record the date and time of the user’s intervention. In addition, Datasets are immutable, and as such all modifications made to the original data must be uploaded in separate Datasets using version numbers. Last, access to the system is restricted to authorized users, who can have different roles.<ref name="Bauch_openBIS11" /> All these features are in accordance to the FDA regulations for electronic records.<ref name="21CFR11@ecfr">{{cite web |url=http://www.ecfr.gov/cgi-bin/retrieveECFR?gp=&SID=04a3cb63d1d72ce40e56ee2e7513cca3&r=PART&n=21y1.0.1.1.8 |title=Electronic Code of Federal Regulations - Title 21: Food and Drugs - Part 11: Electronic Records; Electronic Signatures |publisher=U.S. Government Printing Office |accessdate=25 October 2016}}</ref> | |||
===LIMS=== | |||
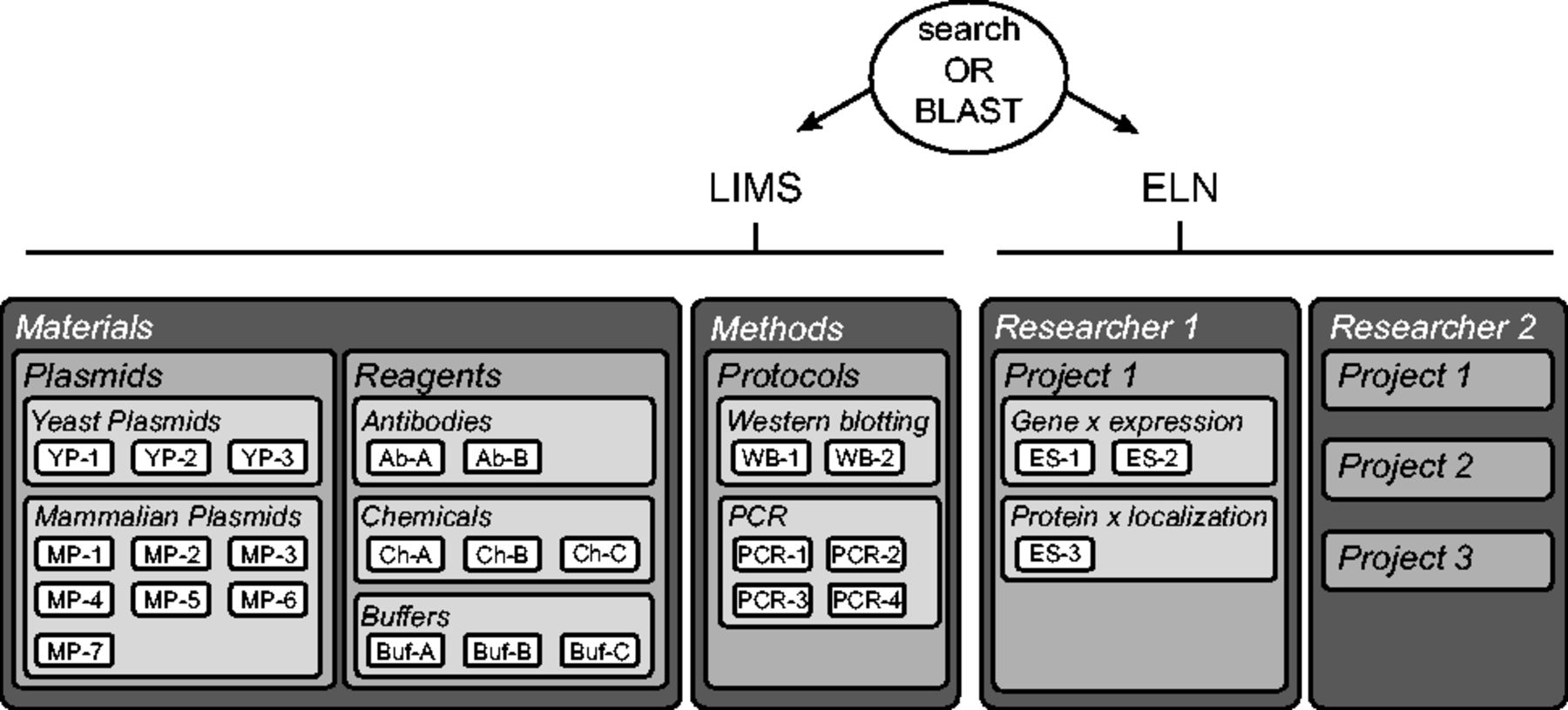
The LIMS consists of two "Data Spaces," the "Materials" and the "Methods," accessible and editable by all group members (Fig. 2 , left side). | |||
[[File:Fig2 Barillari Bioinformatics2015 32-4.jpg|800px]] | |||
{{clear}} | |||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| border="0" cellpadding="5" cellspacing="0" width="800px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 2.''' Overview of a typical openBIS ELN-LIMS platform. The LIMS consists of the Materials and Methods Data Spaces, where the Samples are organized in Projects and Experiments. The ELN can have several Data Spaces assigned to each researcher in the laboratory. Researchers can organize their own Data Space by creating Projects, Experiments and Samples. Each Sample has a unique identifier. For simplicity, Datasets are not illustrated here.</blockquote> | |||
|- | |||
|} | |||
|} | |||
==References== | ==References== | ||
Revision as of 23:01, 25 October 2016
| Full article title | openBIS ELN-LIMS: An open-source database for academic laboratories |
|---|---|
| Journal | Bioinformatics |
| Author(s) | Barillari, Caterina; Ottoz, Diana S.M.; Fuentes-Serna, Juan M.; Ramakrishnan, Chandrasekhar; Rinn, Bernd; Rudolf, Fabian |
| Author affiliation(s) | ETH Zürich, Lifescience Zürich |
| Primary contact | Email: brinn at ethz dot ch -or- fabian dot rudolf at bsse dot ethz dot ch |
| Year published | 2015 |
| Volume and issue | 32 (4) |
| Page(s) | 638-640 |
| DOI | 10.1093/bioinformatics/btv606 |
| ISSN | 1460-2059 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | http://bioinformatics.oxfordjournals.org/content/32/4/638.long |
| Download | http://bioinformatics.oxfordjournals.org/content/32/4/638.full.pdf (PDF) |
Abstract
Summary: The open-source platform openBIS (open Biology Information System) offers an electronic laboratory notebook and a laboratory information management system (ELN-LIMS) solution suitable for the academic life science laboratories. openBIS ELN-LIMS allows researchers to efficiently document their work, to describe materials and methods and to collect raw and analyzed data. The system comes with a user-friendly web interface where data can be added, edited, browsed and searched.
Availability and implementation: The openBIS software, a user guide and a demo instance are available at https://openbis-eln-lims.ethz.ch. The demo instance contains some data from our laboratory as an example to demonstrate the possibilities of the ELN-LIMS (Ottoz et al., 2014). For rapid local testing, a VirtualBox image of the ELN-LIMS is also available.
Introduction
Scientific recording is an essential part of research, as progress in science depends among other factors on existing knowledge and reproducibility. Recording is the process by which the scientific question, the choice of an experimental procedure, materials and methods, data analysis and interpretation of the results are gathered. Traditionally, all these details were kept in paper notebooks.
In today’s academic life science laboratories, most raw data, analyzed data and scripts are stored electronically, while the experimental details are often recorded in paper notebooks. This makes linking and retrieving experimental details and results problematic. Moreover, the large amount of data produced by today’s readout machines adds a level of complexity as the devices the data are stored on might soon be outdated.
Databases represent a solution to store all the necessary information together[1] because (i) they ensure long-lasting data storage by being independent of the operating environment; (ii) they are installed on a dedicated server, enabling long-term data storage and regular backups; (iii) they allow storage of large amounts of data; (iv) they are easily accessible via a local area network or the internet; (v) custom applications can be easily added.
Several commercial database solutions for ELNs are available, but they are not commonly adopted by academia. An internal evaluation among the research groups of the departments of Biosystems Science and Engineering and Biology of ETH Zurich found that the options affordable to academic labs had either unsuitable user interfaces (UI) or lack of features. The combination of an easy-to-use UI, flexible adaptation to a labs needs, support of an audit trail, physical support of the data server, support for managing large amounts of data and for integrating new measurement devices was only available at very high costs. On the other hand, the available open-source platforms lack many of the above features and, as they often originate from a single person within a research group, they also lack maintenance and support during the required data management life cycle.
Here, we describe the open-source database openBIS (open Biology Information System) electronic laboratory notebook and a laboratory information management system (ELN-LIMS), a software specifically developed with academic life science labs in mind. The structure of the database ensures that the results are represented in a logical and meaningful fashion, and it guarantees that the users can find, understand and reuse the data. openBIS ELN-LIMS consists of two interconnected parts: the LIMS, where the information about materials and methods is stored and kept up to date and the ELN, where data obtained from experiments are uploaded and annotated. The system is based on the openBIS platform[2], which, since 2007, is actively developed and maintained by a dedicated team of software engineers at the ETH and is productively used in several academic institutions and a few private companies.
Results
System's overview
Our goal was to implement a database to support the research activity of a typical biology laboratory. We conceived it as a tool to assist everyday activities: storage of materials, methods set-up, design and description of experiments, data labeling, storage and organization of results. To this purpose, we based our system on the platform openBIS[2], as it has the necessary features described above, is field-tested since about seven years and known to work well also with hundreds of terabytes of data.
The openBIS core has a predefined hierarchical structure (Fig. 1). The top level, called "Data Space", controls access and rights and contains one or more "Projects," which in turn contain one or more "Experiments." "Samples" are the basic unit of openBIS and are organized within Experiments. We adapted this hierarchical structure to describe the organization and the objects of a typical life science laboratory. We defined the type and the number of Experiments and Samples and assigned customized properties to describe metadata. External files, in any kind of format, are uploaded as "Datasets" to each Sample. Each Sample can have any number of Datasets. We defined different types of Datasets and defined appropriate sets of properties (or annotations).
|
We used parent–child relationships linking Samples or Datasets to describe the logical or physical connections existing between the corresponding objects in the real world. We introduced the possibility to describe the nature of these relationships by annotating the links using texts or keywords.
To ensure that the creation, modification or deletion of any electronic records is traceable, computer-generated, time-stamped audit trails are used to record the date and time of the user’s intervention. In addition, Datasets are immutable, and as such all modifications made to the original data must be uploaded in separate Datasets using version numbers. Last, access to the system is restricted to authorized users, who can have different roles.[2] All these features are in accordance to the FDA regulations for electronic records.[3]
LIMS
The LIMS consists of two "Data Spaces," the "Materials" and the "Methods," accessible and editable by all group members (Fig. 2 , left side).
|
References
- ↑ Bird, C.L.; Willoughby, C.; Frey, J.G. (2013). "Laboratory notebooks in the digital era: The role of ELNs in record keeping for chemistry and other sciences". Chemical Society Reviews 42 (20): 8157-75. doi:10.1039/c3cs60122f. PMID 23864106.
- ↑ 2.0 2.1 2.2 Bauch, A.; Adamczyk, I.; Buczek, P. et al. (2011). "openBIS: A flexible framework for managing and analyzing complex data in biology research". BMC Bioinformatics 12: 468. doi:10.1186/1471-2105-12-468. PMC PMC3275639. PMID 22151573. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3275639.
- ↑ "Electronic Code of Federal Regulations - Title 21: Food and Drugs - Part 11: Electronic Records; Electronic Signatures". U.S. Government Printing Office. http://www.ecfr.gov/cgi-bin/retrieveECFR?gp=&SID=04a3cb63d1d72ce40e56ee2e7513cca3&r=PART&n=21y1.0.1.1.8. Retrieved 25 October 2016.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added.