Journal:Practical approaches for mining frequent patterns in molecular datasets
| Full article title | Practical approaches for mining frequent patterns in molecular datasets |
|---|---|
| Journal | Bioinformatics and Biology Insights |
| Author(s) | Naulaerts, S.; Moens, S.; Engelen, K.; Vanden Berghe, W.; Goethals, B.; Laukens, K.; Meysman, P. |
| Author affiliation(s) | University of Antwerp, Antwerp University Hospital, Fondazione Edmund Mach |
| Primary contact | Email: pieter dot meysman at uantwerpen dot be |
| Editors | Dandekar, T. |
| Year published | 2016 |
| Volume and issue | 10 |
| Page(s) | 37–47 |
| DOI | 10.4137/BBI.S38419 |
| ISSN | 1177-9322 |
| Distribution license | Creative Commons Attribution 3.0 Unported |
| Website | http://www.la-press.com/ (HTML) |
| Download | http://www.la-press.com/ (PDF) |
Abstract
Pattern detection is an inherent task in the analysis and interpretation of complex and continuously accumulating biological data. Numerous itemset mining algorithms have been developed in the last decade to efficiently detect specific pattern classes in data. Although many of these have proven their value for addressing bioinformatics problems, several factors still slow down promising algorithms from gaining popularity in the life science community. Many of these issues stem from the low user-friendliness of these tools and the complexity of their output, which is often large, static, and consequently hard to interpret. Here, we apply three software implementations on common bioinformatics problems and illustrate some of the advantages and disadvantages of each, as well as inherent pitfalls of biological data mining. Frequent itemset mining exists in many different flavors, and users should decide their software choice based on their research question, programming proficiency, and added value of extra features.
Keywords: frequent itemset mining, protein domain structure, protein–protein interaction, gene expression, Mycobacterium tuberculosis
Introduction
In the last decade, various information-rich resources have become available to study organisms on a systems-wide scale. The rapid accumulation of complex biological data in extensive compendia demands powerful and specialized pattern mining techniques.[1][2][3][4] A popular group of pattern mining techniques are itemset mining and their derivative, association rule mining. These methods are typically known for their ability to detect frequently co-occurring products in lists of customer supermarket baskets, effectively identifying the patterns in customers’ shopping behavior.[5] In this context, the shopping cart is formally known as a transaction, while the individual products are the items. The discovery of sets of correlated items (i.e. itemsets) is the goal of this data mining approach, which can be highly relevant in the context of life sciences. For example, one can investigate which genes are often coexpressed in tissue samples or which mutations often occur together in cancer tumors of a given type.
Frequent itemset mining has proven especially useful in capturing and summarizing the characteristics of complex datasets to their important and most interesting aspects. Frequent patterns can be converted into rules with a discriminatory value that can, in turn, be used to build transparent classifications. For example, if a gene C is always upregulated when genes A and B are downregulated, the frequent itemset {A|Down, B|Down, C|Up} can be rewritten as the rule {A|Down, B|Down} ≥ {C|Up}, where the left-hand side (antecedent) of the rule leads to the consequent (right-hand side) of the rule. Rules of this type can be used to distinguish between tumor types, gene clusters, and various other biological contrasts. The advantage of this approach is that the rules immediately explain why a particular label was given, which is an advantage over machine learning methods such as neural networks that act as a black box. The strengths of frequent itemset mining have been consequently demonstrated in a broad range of bioinformatics applications, ranging from gene expression data[6][7][8], annotation mining[9][10], and combinations thereof[11][12] to interaction networks.[13] A comprehensive overview of the broad range of implementations and bioinformatics applications of frequent itemset mining techniques was recently published.[14]
Despite their demonstrated suitability to address various bioinformatics problems, frequent itemset mining techniques have not been generally adopted in day-to-day omics data analysis workflows, and their popularity is only slowly gaining traction. This can be partially attributed to a number of shortcomings in the existing implementations. First of all, most are command line tools that often need to be compiled from the source code, and clear documentation regarding their installation is often lacking. This lack of user-friendliness poses a serious entrance barrier that daunts many life scientists. Second, the output of the implementations is often presented in a format that is not readily interpretable by domain experts. The results of the mining process are typically long pattern lists containing flat text files. However, these lists are often very lengthy and highly redundant. This is caused, in part, by the fact that if a set is frequent, any of the smaller subsets that it contains will also be frequent. This is also known as the apriori principle. For many pattern mining applications, there is often a so-called pattern explosion with results that list millions of patterns. Due to the verbose nature of these lists, user-friendly tools to process, query, and visualize this output are indispensible.
Convenient prioritization, filtering, cleaning, and interpretation of pattern result lists require certain functionalities that are rarely covered by existing implementations. Third, iterative optimization of the pattern list and browsing through the output of these algorithms is often hard, as they create static output that needs to be processed and converted to a compatible format before the next step in the iterative mining process can start. This can make result prioritization, an inherent part of many pattern discovery projects, a very cumbersome process.[14]
To address some of these limitations, software frameworks have been developed for interactive visual pattern mining, such as the MIME tool.[15] Such toolboxes offer intuitive access to interestingness measures, mining algorithms, and post-processing algorithms to assist in identifying interesting patterns. By enabling interactive mining, it allows the user to combine their subjective interestingness measure and background knowledge with a wide variety of objective measures to easily and quickly mine the most important and interesting patterns. In this article, we demonstrate the opportunities of frequent itemset mining in real-world bioinformatics scenarios and describe the application of three commonly used methods, namely, Apriori[5], arules[16], and MIME.[15] This comparison is based on three representative bioinformatics use cases, i.e. domain co-occurrence within proteins, interactions between domains in interacting proteins, and the response of the pathogen Mycobacterium tuberculosis to several drug treatments. For this purpose, we utilize data from UniProt[2], IntAct[4], and COLOMBOS.[1] The data files and step-by-step tutorials on how to install and run the three presented tools on the three use cases are available in Supplementary Files 1–5. The goal of this study is to explore how interesting and biologically relevant patterns can be effectively generated with different tools and provide the community with some guidance on how frequent itemset mining tools can be used in complex life science scenarios.
Materials and methods
Frequent itemset mining
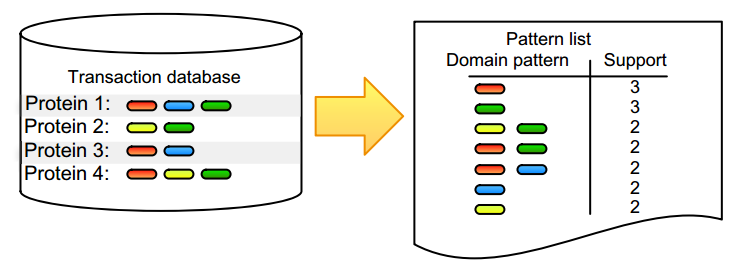
For datasets with large amounts of objects, features, or observations, it is not tractable to check all possible combinations and find correlated annotation terms or biological entities. Frequent itemset mining is composed of a set of tools that are able to find co-occurring terms, known as items, in big data. These items can be any entity, ranging from genes and RNAs to proteins or drugs, and thus allow, for example, to identify co-expressed genes or proteins. The mining algorithms typically start from transactional databases, as shown in Figure 1.
|
A transaction is simply a set of items, and a transactional database layout contains one transaction per row, in which each transaction refers to an observation, such as the collection of domains associated with a protein (Fig. 1) or the aggregation of supermarket articles found in a shopping basket that was checked out by a single customer. These algorithms use heuristics to reduce the search space. For example, priori-based methods use the Apriori principle, which states that if an itemset is not frequent, all supersets of these items will also not be frequent. A pattern is frequent when its items appear more than expected, which means that its support, the absolute number of times that a pattern occurs, needs to exceed a userdefined minimal threshold. It is worth noting that frequency and support are often inter-exchanged, with frequency thresholds stating the relative minimum (often 10%) threshold that needs to be exceeded. A hot topic in this field is the inability of frequent itemset mining to identify co-occurrence of continuous values, as most methods require the user to conduct well-chosen discretization steps that define the items evaluated by the algorithm. We have described our discretization steps where needed in the case studies described subsequently.
Datasets
In order to compare the benefits of each frequent itemset method, several datasets were created from public resources, more specifically using InterPro, COLOMBOS, and IntAct. For each of the case studies, a transaction dataset was constructed based on the data downloaded from the database and saved in a simple space- or tab-delimited file. All of the transaction data files have been made available in Supplementary Files 1–3. A tutorial on how to practically execute the mining process has also been included (Supplementary File 4), as well as a small Python script that compares the mining results for the Borgelt’s Apriori implementation (Supplementary File 5).
Protein domain analysis
For the first use case, we downloaded all known proteins of the human reference proteome on February 19, 2014 from UniProt[2] and retained only those that contained at least one InterPro domain.[17] In total, this resulted in a set consisting of 20,636 proteins. In the second use case, we mapped this information on top of the IntAct protein–protein network[4], which was downloaded on the same day.
COLOMBOS
All the microarray information was obtained from COLOMBOS (a collection of microarrays for bacterial organisms)[1], which contains numerous renormalized gene expression experiments extracted from the Gene Expression Omnibus[18] and ArrayExpress.[19] Using the advanced search option, we created a dataset for M. tuberculosis composed of several experiments in which the bacteria responded to antibiotics added to their growth medium. Next, the information in this dataset was discretized based on the fold change. Traditionally, log2-fold changes ranging from 1 to 1.5 are used to define the differential regulatory state of a gene. Here, we used a log2-fold change of 1.2, which results in a dataset that includes 25% of the protein-coding genes in M. tuberculosis that were differentially expressed in at least one condition contrast. All log2-fold changes greater than 1.2 were considered upregulated, while those smaller than −1.2 were labeled as being downregulated. Fold changes between these two values were excluded from the dataset. For each gene, the log-fold change was simplified to a discretized state (up or down) and appended as a suffix to the contrast information. For example, the gene Rv0823c has been identified to be downregulated (fold change −1.43) in study GSE1642, in which the treatment is an addition of 1 µM valinomycin. The combination of all this information into one label results in Rv0823c|Down, which is a discrete item that can be used in the mining process.
Tools
Apriori
Apriori is one of the oldest, most simple, and popular frequent itemset mining algorithms.[20] It is available in various implementations that vary from command line tools to parts of data analysis software suites. In this article, we use Christian Borgelt's Apriori implementation, which is available at http://www.borgelt.net/apriori.html.[21] We compiled the C source code and ran the implementation using the synthaxis./apriori [options] infile [outfile] from the terminal on a Macintosh running OS 10.9 Mavericks.
One of the major advantages of Borgelt’s Apriori version, other than its improved pattern identification efficiency, is its support for distinctly different types of itemsets, including maximal, closed, and open itemsets. By default, Apriori generates all possible itemsets (open), which are typically far too many to analyze. The information contained by these patterns is largely redundant, which means that the resulting pattern list can be efficiently reduced with a minimal loss of information content. For example, only itemsets that have no frequent superset can be retained (maximal itemsets). This results in a major reduction in patterns that cannot be justified in every scenario. A more balanced general approach that still effectively reduces the output of the mining process consists of mining only closed patterns. These are formally defined as itemsets that have no immediate superset with the same support value. Although all these three pattern classes have been used to approach various life science problems, closed itemsets have been the most prominent in analyses that deal with the typical genome- or proteome-wide scale datasets.[14]
arules
In addition to command line tools, several efforts have been made to bring frequent itemset mining to other platforms. One such project is the R package arules, which also contains the Apriori implementation by Borgelt in addition to other mining algorithms.[16]
arules provides an entire toolbox for the representation, manipulation, and analysis of frequent itemsets and association rules in R. It contains several scoring metrics and allows the calculation of various properties, such as dissimilarity. arules is also compatible with arulesViz[22], which allows rapid visualization of the mining results. arules is available for download through the R interface as a CRAN distribution.
MIME
The third tool covered in this study is MIME.[15] MIME provides a dynamic and interactive graphical environment to interact with the dataset and retrieve patterns. It hosts several popular pattern mining algorithms, such as Apriori[5], Eclat[23], tiling[24], top-K mining[25], the OPUS Miner[26], and Carpenter[6], as well as various classification algorithms that are based on association rule mining.[27][28][29] Similar to arules, MIME has various scoring measures (so-called interestingness metrics) and, furthermore, supports iterative mining. The software is written in Java and depends on two Java libraries, namely, QTJambi (http://qt-jambi.org) and WEKA[30], and is available from http://adrem.ua.ac.be/mime.
Results and discussion
By means of practical use cases, we will go through increasingly complex life sciences scenarios that at each step require additional preprocessing steps to translate the problem into a format recognizable by the tools.
Case study 1: Analyzing domain co-occurrence within proteins
The simplest use case for frequent itemset mining is the analysis of domain co-occurrence within proteins. The transaction dataset contains all human proteins documented in InterPro with at least one protein domain. Herein, each protein is considered as a transaction, which equals a single line in the input file containing the domains (items). In total, 20,636 proteins were present in the dataset. All three tools were able to find the most frequent individual domains by setting the maximal number of items in an itemset to one. MIME delivers this information without requiring an extra step. This step allows us to identify which domains occur the most in the dataset and which is important for the calculation of several quality measures, such as lift value. It also gives the researcher a quick initial idea of which domains are most likely to appear in frequent itemsets. If these domains associate in an aspecific manner with another domain and can be expected by random combination (low lift value), the pattern is not interesting for a biologist who tries to identify domains that are functionally correlated.
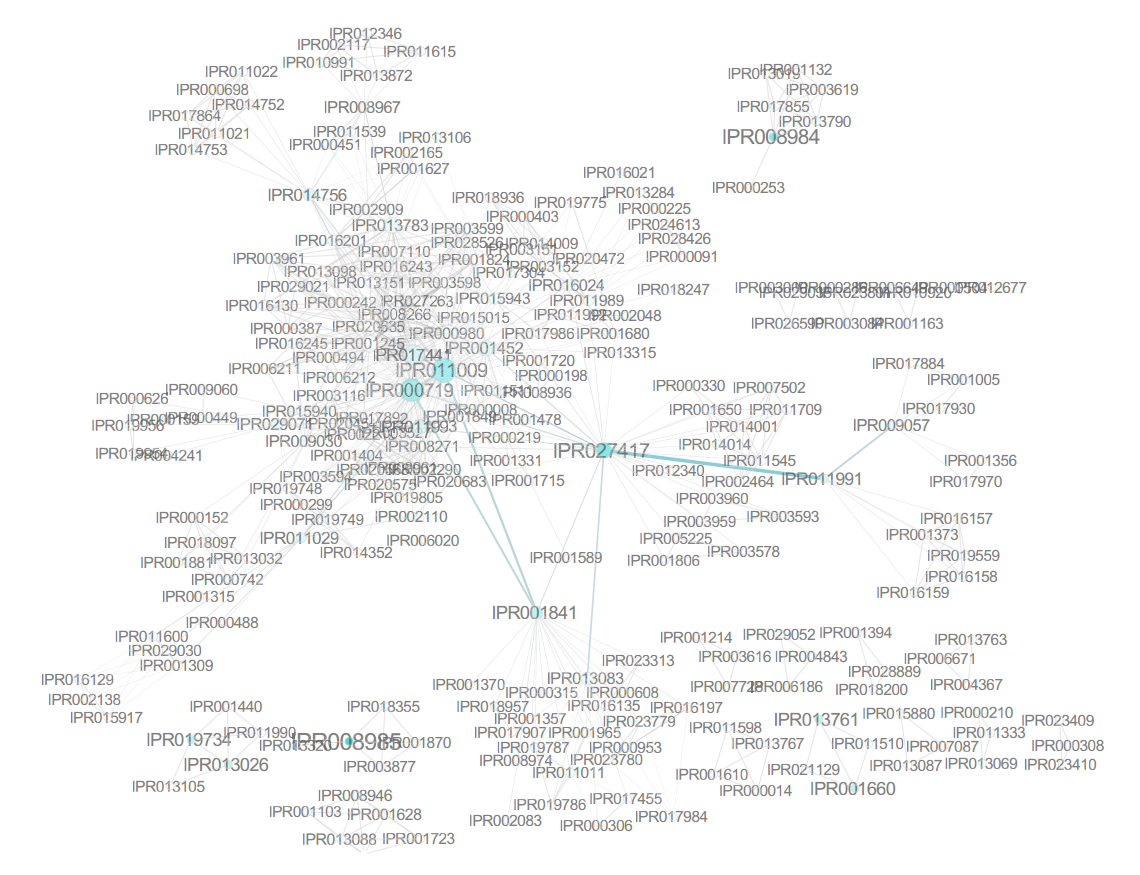
The most frequent domain was identified to be IPR027417, a P-loop containing nucleoside triphosphate hydrolase, which occurred 1329 times (6.44% of all proteins). Other than this domain, only IPR013783 (4.44%), IPR011009 (4.31%), IPR000719 (3.96%), and IPR015943 (3.19%) occur in more than 3% of the human proteins. IPR000719 and IPR011009 (protein kinase-like domain) occurred together in the majority of proteins they occurred in, as represented by the corresponding support value (3.94%). This pattern was the most frequent of all associated InterPro terms, with the highest support and a high lift value, which suggests a nonrandom association. However, this can easily be attributed to the fact that the former is a protein kinase domain and the latter is a protein kinase-like domain, with IPR000719 being a subset of IPR011009 and kinases being a family containing a significant number of proteins. Interestingly, one would then expect IPR000719 (819 proteins) to always co-occur with IPR011009, but this was apparently not the case for five proteins, namely, C9JE15 (aarF domain-containing protein kinase 2), D6RHX9 (calcium/calmodulin-dependent protein kinase type II subunit alpha), E9PPN3 (N-terminal kinase-like protein), F8W0N2 (serine/threonine-protein kinase receptor R3), and H0YAH6 (epithelial discoidin domain-containing receptor 1). In the following updates of UniProt, these five proteins had the IPR011009 domain added, which illustrates that even basic frequent itemset mining can be applied to detect inconsistencies in annotations.
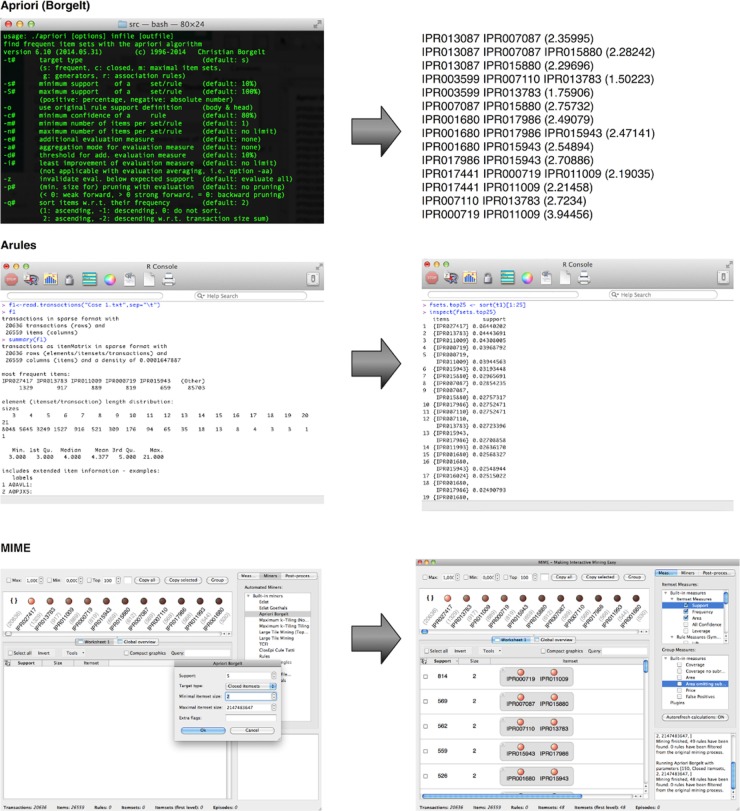
A combination of the low cutoff percentages and the complexity of biological data make setting a minimal threshold a rather daunting task. One approach is iteratively lowering the threshold and keeping it above the value where the number of patterns explodes. This parameter fine-tuning may be time consuming, and the arbitrary threshold can be hard to justify biologically. In this case, it can be useful to simply search for 100 most frequent patterns without much knowledge of their individual protein abundance. In many settings, the manual evaluation of the pattern results will limit itself to those patterns that have the highest support value, as these will be most frequent in the data set and often the most relevant or interesting. For such a purpose, top-K mining would be much more straightforward as the only parameter it requires is setting K to 100. Of the three implementations addressed, this is only possible with MIME. Top-K mining with the other solutions requires knowledge of Java or can be cumbersome.[31][32] Figure 2 shows the output of the three itemset mining implementations. For Apriori and arules, we iteratively optimized a lower threshold in order to display these results. An overview of the most frequent itemsets obtained with MIME is listed in Table 1.
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
The 100 most frequent itemsets feature combinations of 63 unique protein domains. IPR013783 (immunoglobulinlike fold), IPR001881 (epidermal growth factor [EGF]-like calcium binding domain), IPR000152 (EGF-like aspartate/ asparagine hydroxylation site), IPR018097 (EGF-like calcium binding), IPR013032 (EGF-like conserved site), and IPR000742 (EGF-like domain) appear in over 11 unique patterns of these 100 most frequent itemsets.
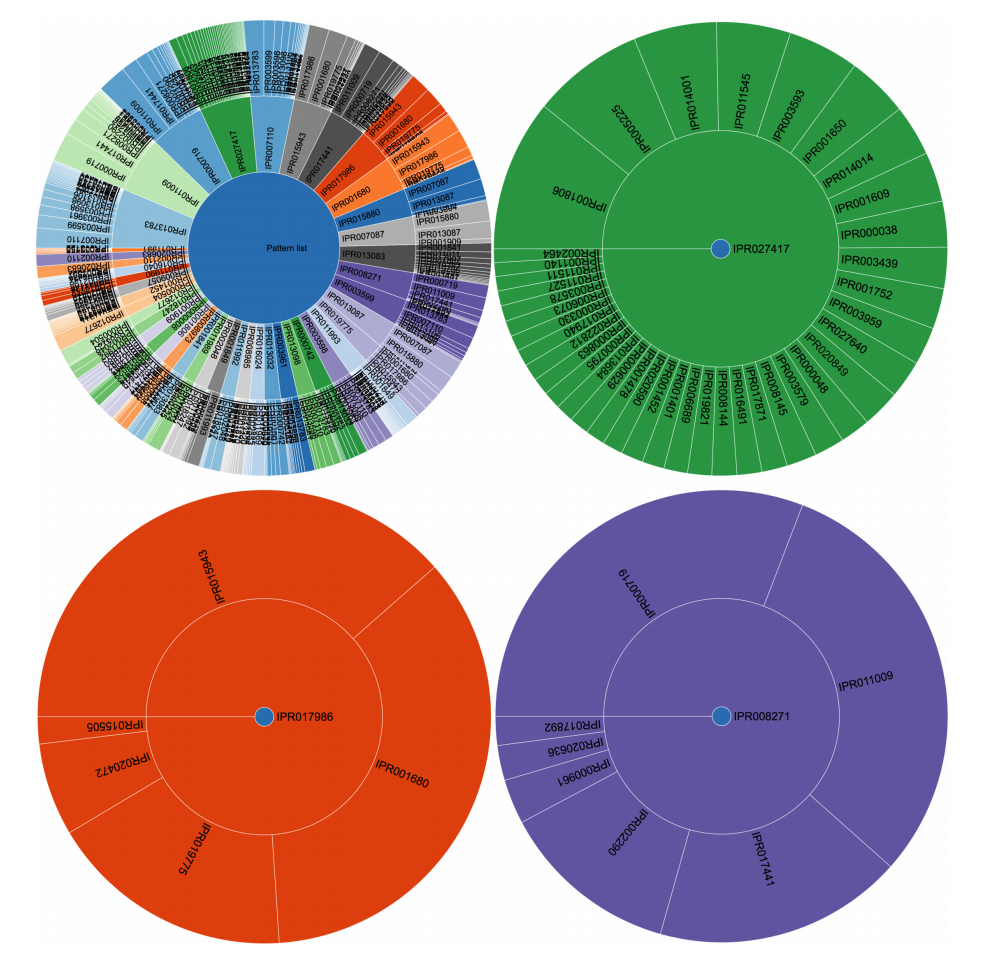
The use of frequent itemset mining techniques, without accounting for the hierarchical structure of annotations, will typically lead to this kind of frequent but trivial patterns with no true informative value. For example, IPR007087 (zinc finger, C2H2) is a child of the CH2-like zinc fingers (IPR015880). Patterns of this kind create additional overhead that needs to be filtered out to separate it from patterns with true informative value. This problem has already been elaborately described in literature, especially for Gene Ontology analysis.[10][33] Therefore, we grouped domains into functional categories, based on the structure of the InterPro annotation tree, to remove the redundant annotations and found immunoglobulin-like domains, CH2 zinc fingers, protein kinase-like domains, and WD40 repeats to be the most prevalent in human proteins when looking at individual domains. However, the 100 most frequent itemsets then consisted of vastly different frequent patterns at much lower frequencies. For example, some of the most prevalent patterns were the co-occurrence of protein kinaselike domains (IPR011009) and the ATP-binding domain (3.14%). Co-occurrence within tyrosine kinases and SH2- domains was also prevalent. A full overview of the 100 most frequent itemsets is shown in Supplementary File 6. Overall, frequent itemset mining can be used to cluster proteins based on their domain annotations and explore the structure and relationships that may exist in this dataset. Figure 3 shows the mined patterns in a pie chart representation, from three broad categories that exist in the dataset, namely, the kinase-related, helicase-related, and WD40-related patterns. Each pattern in Figure 3 is represented as a path from the center to the exterior rings. Neither tool required extensive programming skills to tackle this use case.
|
Case study 2: Mining for co-occurring domains between interacting proteins
A topic of great biological interest is the identification domains that are likely to play key roles in protein–protein interactions, in, for example, drug discovery.[34] We mapped the InterPro domains[17] corresponding to each UniProt identifier[2] in the human IntAct protein–protein interaction network[4] into transactions. Each transaction then represents a pair of proteins, and the items within a transaction correspond to the union of domains for both proteins in the pair. We then removed all excess protein domains using the grouping method described in case 1 to prevent noninformative patterns from hierarchical annotations from appearing. However some patterns may still result from intraprotein domain co-occurrences, as we did not consider the protein of origin for each of the domains. Therefore, the intraprotein patterns obtained in case 1 were subtracted from the combined list. For Borgelt's Apriori implementation, we had to perform the mining process for both datasets using the lowest possible threshold to avoid the pattern explosion and then removed all the patterns appearing in the list intersection with a Python script. In arules, the results of both mining processes could be compared using data frames within the R framework[35], with a basic understanding of R language syntax. MIME required no such scripting effort and allowed to compare the result files from the two mining processes directly using the "compare worksheets" function.
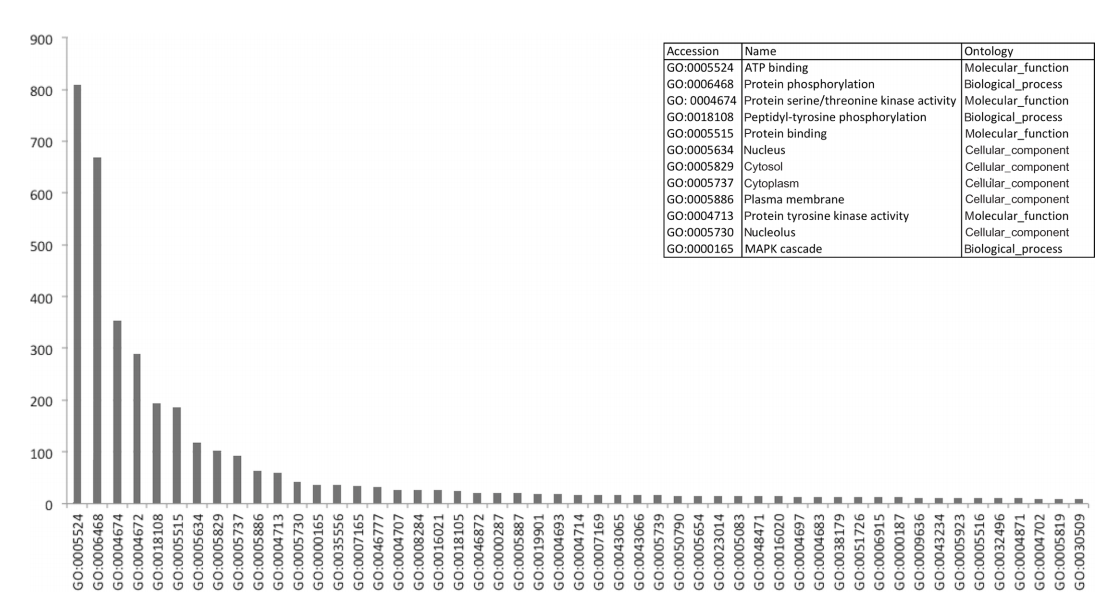
Either tool was able to find large amounts of frequently recurring motifs within proteins that have been described in literature, with notable examples such as interactions between the 14-3-3 domain (IPR000308) and S/T protein kinase domain (IPR002290).[36] In the IntAct network, we identified a total of 272 interactions that exhibited this particular pattern, with a total of 76 unique proteins being involved (42 kinases). Figure 4 shows the distribution of the functional annotations of the proteins corresponding to the domains in the interaction dataset. As can be seen in Figure 4, kinase interactions seem to be the most common in the human interactive dataset, with all of the top functions referring to this regulatory mechanism. An overview of the 100 most frequent patterns is shown in Supplementary File 7. Frequent itemsets may also be used to detect complexes, as indicated by the domain associations between Skp1-containing proteins and F-box proteins, which have been previously described.[37][38]
|
We grouped all the proteins corresponding to a rule and looked for potential biological enrichments using classic overrepresentation based on hypergeometric testing with Benjamini–Hochberg correction (P-value , 0.05). The results, visualized in Cytoscape[39], are shown in Figure 5. There is a clear functional coherence in interacting proteins, which is naturally reflected in the combination of elements at the domain level. For example, many proteins involved in apoptosis were characterized as containing the death-like domain (IPR011029) and binding sites for TNF (IPR006035) and formed a more densely interconnected protein network with their associated kinases.
|
Compared with the textual output of Apriori and arules, navigating through the dataset using MIME is more flexible. For example, a domain of interest could be dragged into the pattern browser window and extended by adding other domains in a drag and drop fashion. MIME automatically recalculates all quality measures it contains each time a modification is made to the pattern list. This means that the lift values, area, coverage, support, and many more measures are consistently up to date. This enables a more dynamic approach to manual supervision of the pattern list, which can speed up the discovery of interesting properties in a dataset.
It should be mentioned that specialized frequent itemset mining approaches exist to rapidly find patterns that discriminate between sets or more formally defined, i.e. patterns whose support value increase significantly from one dataset compared with the other. Discriminative or emergent patterns have been suggested to have a higher value for use in classification models and have been extensively reviewed.[40] In this case study, such methods could be used to more rapidly identify the patterns that are more likely to be the result of interacting proteins or, by extension, distinguish between protein families.
Case study 3: M. tuberculosis response to stress
In the third case study, we downloaded gene expression profiles of M. tuberculosis in several antibiotic studies from the microbial gene expression compendium COLOMBOS.[1] We discretized the fold changes of the expression into upregulated (fold change >1.2) and downregulated (fold change <−1.2). Discretization is essential for traditional frequent itemset mining. In this case, the transactions consist of the different expression experimental contrasts, and their items are the up- or downregulated genes within these contrasts. In total, the dataset contains 47 contrasts, and the most frequent differentially expressed items were WhiB7|Down (21% or 47%), esxS|Up (16% or 34%), higB|Down (16% or 34%), PE20|Down (16% or 34%), and esxH|Up (15% or 32%). Supplementary File 8 shows the resulting mining output.
The most frequent itemsets have a support value of 14, and several underlying types of frequent itemsets are immediately apparent, either in the graphical display of MIME or in the textual output of arules and Apriori. A first type of pattern refers to the nature of transcription in prokaryotes. For example, esxH|Up (16) and esxS|Up (15) were strongly correlated in an itemset with a frequency of 30% (14). In the two cases, both genes did not occur together, due to marginally falling outside the boundaries set by the discretization procedure. esxH and esxS are part of the same transcription unit, which codes a Type VII secreted compound (EsxH) that impairs trafficking in the host organism.[41] In order to fulfill its purpose, it acts in concert with esxG, which is in the same transcription unit and thus also upregulated. esxG appeared nine times in our dataset, each time upregulated (7) or downregulated (2) when the other elements of the operon were as well.
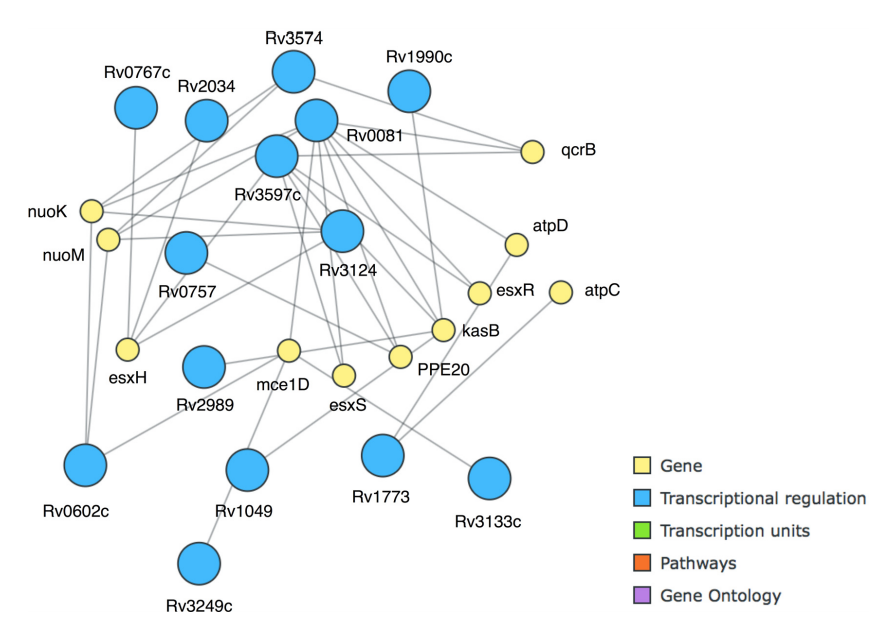
Another key pattern turned out to be PE20|Down and whiB7|Down, also with 14 as the support value. WhiB7 is a transcriptional activator important for antibiotic resistance[42], and PE20 is a largely uncharacterized protein with a length of 99 amino acids that has an effect on pathogenicity[43], but their support value, in comparison with the large amount of rules these two genes appear in, suggests a key role in M. tuberculosis response to several antibiotics. This pattern could be further extended to include higB|Down, esxH|Up, esxR|Up, and kasB|Up with a support value of 10 (21%). This was especially easy to do using MIME and the "best extension of basket" functionality, which greatly facilitates pattern exploration with the otherwise often laborious task of browsing through the often vast sets of patterns originating from more extensive datasets. We extended the itemset to the maximal length at a minimal support value of 7, resulting in a set of 11 genes enriched in the generation of precursor metabolites and energy metabolism (GO:0006091) that is strongly interconnected at transcriptional levels (Fig. 6).
|
We then investigated which antibiotic treatments contributed to it, as seven transactions supported it (and thus maximally seven different drug treatments). We found these contrasts to be Streptomycin:5 (GSE1642), Amikacin:10 (GSE1642), Streptomycin:5 (GSE1642), Amikacin:5 (GSE1642), Amikacin:5 (GSE1642), Tetracycline:10 (GSE1642), and Capreomycin:10 (GSE1642), respectively. Amikacin, capreomycin, and streptomycin are aminoglycoside antibiotics that tend to target the initiation of protein synthesis by causing mistranslations.[44] Interestingly, tetracycline, which acts by disruption of normal mRNA recognition by the tRNA anti-codon, was associated with the very same itemset even though the antibiotic has a vastly different structure and mode of action. As such, frequent itemsets that combine essential disturbances of genes may prove interesting to identify novel compounds that are vastly different, but exhibit a similar downstream effect on gene expression. Exploratory analysis with user-friendly pattern mining software can be a great assistance for this task.
Conclusions
Several implementations, which do not require extensive programming skills, exist to perform biological data mining with frequent itemsets. In three typical bioinformatics experiments, we found each method to excel at different aspects. Borgelt’s Apriori implementation was very fast using the command line, but lacked the flexibility that arules has in the R environment. However, arules requires the user to have an understanding of the R scripting environment, and this is characterized by a steeply increasing learning curve for more complex operations, such as comparing results from two mining processes. MIME showed its value when subsequent mining steps were required, as the output of one mining step did not have to be processed to be usable in the next mining iteration. This is especially useful in exploratory analysis of complex biological datasets. The presence of additional mining algorithms in a graphical environment in which patterns can be modified by simply dragging and dropping items further strengthens its ease of use.
Author contributions
Conceived and designed the experiments: SN, SM, BG, KL. Analyzed the data: SN, KE, WVB, KL, PM. Wrote the first draft of the manuscript: SN, PM. Contributed to the writing of the manuscript: SN, SM, KE, WVB, BG, KL, PM. Agreed with manuscript results and conclusions: SN, SM, KE, WVB, BG, KL, PM. Jointly developed the structure and arguments for the paper: SN, KL, PM. Made critical revisions and approved the final version: SN, SM, KE, WVB, BG, KL, PM. All the authors reviewed and approved the final manuscript.
Supplementary materials
Link to .zip file containing all supplementary files: Download here
Description of supplementary materials
Supplementary File 1. Intra-protein dataset. This file contains all transactions containing protein domain presence that was extracted from the InterPro database. Each protein (shown before semicolon) and its transaction (after colon) are listed to make retracing patterns easier. To use in frequent itemset mining, strip everything before the transactions.
Supplementary File 2. Protein domains in binary interaction dataset. This file contains all InterPro domains that were mapped to interactions that were extracted from IntAct. The UniProt accessions of both interaction partners in the binary interaction are shown before the colon. Strip this part to only retain the interactions. Making the contrast between the mining results of Supplementary File 1 and those in this file will remove the protein-specific patterns, as well as several patterns that result from the ontology itself.
Supplementary File 3. M. tuberculosis dataset. This file contains all transactions extracted from the COLOMBOS dataset. Each row represents the response to an antibiotic, in which only the genes that were significantly up- or downregulated according to the logFC discretization step were retained. Each item combines the gene name with its individual response.
Supplementary File 4. Tutorial. This brief tutorial describes the practical use of each of the three frequent itemset implementations on the case studies featured in this article.
Supplementary File 5. Python script. This file contains a simple Python script that compares the output of the Apriori Borgelt implementation to another file with similar structure. This can be used to find overlaps and differences between the mining output of Case 1 and Case 2.
Supplementary File 6. 100 most frequent InterPro intraprotein patterns. The 100 most frequent intra-protein itemsets that contain domain associations. Most itemsets consist of combinations of kinases, patterns resulting from the ontology tree, immunoglobulins, WD40 repeats and others. This file extends Table 1.
Supplementary File 7. 100 most frequent InterPro interprotein domain patterns. This table shows the 100 most frequent domain associations in protein-protein interaction data. We obtained this output by subtracting the intra-protein patterns. We find that the top 100 length 2 rules mostly consist of combinations of only 45 distinct domains. Most of these terms refer to kinase cascades (associations between serine/threonine and tyrosine kinases), interactions between SH2 and SH3 domains and kinases, as well as a large amount of domains that is involved in ubiquitinylation and immunological response. All patterns that could be produced by the InterPro ontology were filtered out, which leaves the interaction between the SH2 and the SH3 domain as the most abundant.
Supplementary File 8. 100 most frequent itemsets in the M. tuberculosis use case. In this table the 100 itemsets are shown that represent the gene regulation response to several antibiotics included in COLOMBOS. Every item is a combination of a gene name and its regulatory state, which was the result of a discretization step (logFC .1.2: upregulated, ,−1.2: downregulated. Several genes, such as whiB7, higB, and PE20, are strongly co-regulated. We were also able to identify transcriptional units, such as the esx-unit.
References
- ↑ 1.0 1.1 1.2 1.3 Meysman, P.; Sonego, P.; Bianco, L. et al. (2014). "COLOMBOS v2.0: An ever expanding collection of bacterial expression compendia". Nucleic Acids Research 42 (D1): D649-D653. doi:10.1093/nar/gkt1086. PMC PMC3965013. PMID 24214998. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3965013.
- ↑ 2.0 2.1 2.2 2.3 UniProt Consortium (2013). "Update on activities at the Universal Protein Resource (UniProt) in 2013". Nucleic Acids Research 41 (D1): D43-7. doi:10.1093/nar/gks1068. PMC PMC3531094. PMID 23161681. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3531094.
- ↑ Maietta, P.; Lopez, G.; Carro, A. et al. (2014). "FireDB: A compendium of biological and pharmacologically relevant ligands". Nucleic Acids Research 42 (D1): D267-72. doi:10.1093/nar/gkt1127. PMC PMC3965074. PMID 24243844. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3965074.
- ↑ 4.0 4.1 4.2 4.3 Orchard, S.; Ammari, M.; Aranda, B. et al. (2014). "The MIntAct project: IntAct as a common curation platform for 11 molecular interaction databases". Nucleic Acids Research 42 (D1): D358-63. doi:10.1093/nar/gkt1115. PMC PMC3965093. PMID 24234451. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3965093.
- ↑ 5.0 5.1 5.2 Agrawal, R.; Imielinksi, T.; Swami, A. (1993). "Mining association rules between sets of items in large database". Proceedings of the 1993 ACM SIGMOD International Conference on Management of Data 22 (2): 207–16.
- ↑ 6.0 6.1 Pan, F.; Cong, G.; Tung, A.K.H. et al. (2003). "Carpenter: Finding closed patterns in long biological datasets". Proceedings of the Ninth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining: 637–642. doi:10.1145/956750.956832.
- ↑ Cong, G.; Tan, K.-L.; Tung, A.K.H.; Xu, X. (2005). "Mining top-K covering rule groups for gene expression data". Proceedings of the 2005 ACM SIGMOD International Conference on Management of Data: 670-681. doi:10.1145/1066157.1066234.
- ↑ Gouda, K.; Zaki, M.J. (2005). "GenMax: An efficient algorithm for mining maximal frequent itemsets". Data Mining and Knowledge Discovery 11 (3): 223-242. doi:10.1007/s10618-005-0002-x.
- ↑ Artamonova, I.I.; Frishman, G.; Gelfand, M.S; Frishman, D. (2005). "Mining sequence annotation databanks for association patterns". Bioinformatics 21 (Suppl 3): iii49-iii57. doi:10.1093/bioinformatics/bti1206. PMID 16306393.
- ↑ 10.0 10.1 Manda, P.; Ozkan, S.; Wang, H. et al. (2012). "Cross-ontology multi-level association rule mining in the gene ontology". PLoS One 7 (10): e47411. doi:10.1371/journal.pone.0047411. PMC PMC3470562. PMID 23071802. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3470562.
- ↑ Martinez, R.; Pasquier, N.; Pasquier, C. (2009). "Mining Association Rule Bases from Integrated Genomic Data and Annotations". In Masulli, F.; Tagliaferri, R.; Verkhivker, G.M.. Computational Intelligence Methods for Bioinformatics and Biostatistics. Springer Berlin Heidelberg. pp. 78–90. doi:10.1007/978-3-642-02504-4_7. ISBN 9783642025044.
- ↑ Tseng, V.S.; Yu, H.-H.; Yang, S.-C. (2009). "Efficient mining of multilevel gene association rules from microarray and gene ontology". Information Systems Frontiers 11 (4): 433-447. doi:10.1007/s10796-009-9156-1.
- ↑ Karpinets, T.V.; Park, B.H.; Uberbacher, E.C. (2012). "Analyzing large biological datasets with association networks". Nucleic Acids Research 40 (17): e131. doi:10.1093/nar/gks403. PMC PMC3458522. PMID 22638576. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3458522.
- ↑ 14.0 14.1 14.2 Naulaerts, S.; Meysman, P.; Bittremieux, W. et al. (2015). "A primer to frequent itemset mining for bioinformatics". Briefings in Bioinformatics 16 (2): 216–31. doi:10.1093/bib/bbt074. PMC PMC4364064. PMID 24162173. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4364064.
- ↑ 15.0 15.1 15.2 Goethals, B.; Moens, S.; Vreeken, J. (2011). "MIME: A framework for interactive visual pattern mining". Proceedings of the 17th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining: 757–760. doi:10.1145/2020408.2020529.
- ↑ 16.0 16.1 Hahsler, M.; Grün, B.; Hornik, K. (2005). "arules – A computational environment for mining association rules and frequent item sets". Journal of Statistical Software 14 (15): 1–25. doi:10.18637/jss.v014.i15.
- ↑ 17.0 17.1 Hunter, S.; Jones, P.; Mitchell, A. et al. (2012). "InterPro in 2011: New developments in the family and domain prediction database". Nucleic Acids Research 40 (D1): D306-12. doi:10.1093/nar/gkr948. PMC PMC3245097. PMID 22096229. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3245097.
- ↑ Barrett, T.; Wilhite, S.E.; Ledoux, P. et al. (2013). "NCBI GEO: Archive for functional genomics data sets - Update". Nucleic Acids Research 41 (D1): D991-5. doi:10.1093/nar/gks1193. PMC PMC3531084. PMID 23193258. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3531084.
- ↑ Rustici, G.; Kolesnikov, N.; Brandizi, M. et al. (2013). "ArrayExpress update - Trends in database growth and links to data analysis tools". Nucleic Acids Research 41 (D1): D987-90. doi:10.1093/nar/gks1174. PMC PMC3531147. PMID 23193272. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3531147.
- ↑ Agrawal, R.; Srikant, R. (1994). "Fast algorithms for mining association rules in large databases". Proceedings of the 20th International Conference on Very Large Data Bases: 487–499. http://dl.acm.org/citation.cfm?id=672836.
- ↑ Borgelt, C. (2003). "Efficient implementations of Apriori and Eclat". Proceedings for the 1st IEEE ICDM Workshop on Frequent Item Set Mining Implementations 2003: 90.
- ↑ Hahsler, M.; Chelluboina, S.; Hornik, K.; Buchta, C. (2011). "The arules R-Package ecosystem: Analyzing interesting patterns from large transaction data sets". The Journal of Machine Learning Research 12 (2): 2021-2025. http://dl.acm.org/citation.cfm?id=2021064.
- ↑ Zaki, M.J.; Parthasarathy, S.; Ogihara, M.; Wei, L. (1997). "New algorithms for fast discovery of association rules". 3rd International Conference on Knowledge Discovery and Data Mining: 283–286. http://dl.acm.org/citation.cfm?id=2021064.
- ↑ Geerts, F.; Goethals, B.; Mielikäinen, T. (2004). "Tiling Databases". In Suzuki, E.; Arikawa, S.. Discovery Science. Springer Berlin Heidelberg. pp. 278-289. doi:10.1007/978-3-540-30214-8_22. ISBN 9783540302148.
- ↑ Han, J.; Wang, J.; Lu, Y.; Tzvetkov (2002). [http://dl.acm.org/citation.cfm?id=844747 "Mining top-k frequent closed patterns without minimum support"]. Proceedings of the 2002 IEEE International Conference on Data Mining: 211–218. http://dl.acm.org/citation.cfm?id=844747.
- ↑ Webb, G.I. (1995). "OPUS: An efficient admissible algorithm for unordered search". Journal of Artificial Intelligence Research 3 (1): 431–465. http://dl.acm.org/citation.cfm?id=1622635.
- ↑ Liu, B.; Hsu, W.; Ma, Y. (1998). "Integrating classification and association rule mining". Proceedings of the 4th International Conference on Knowledge Discovery and Data Mining: 80–86.
- ↑ Li, W.; Han, J.; Pei, J. (2001). "CMAR: Accurate and efficient classification based on multiple class-association rules". Proceedings of the 2001 IEEE International Conference on Data Mining: 369-376. http://dl.acm.org/citation.cfm?id=657866.
- ↑ Yin, X.; Han, J. (2003). "CPAR: Classification based on predictive association rules". Proceedings of the SIAM International Conference on Data Mining: 331-335.
- ↑ Hall, M.; Frank, E.; Holmes, G. et al. (2009). "The WEKA data mining software: An update". ACM SIGKDD Explorations Newsletter 11 (1): 10-18. doi:10.1145/1656274.1656278.
- ↑ Liu, Y.C.; Cheng, C.P.; Tseng, V.S. (2013). "Mining differential top-k co-expression patterns from time course comparative gene expression datasets". BMC Bioinformatics 14: 230. doi:10.1186/1471-2105-14-230. PMC PMC3751367. PMID 23870110. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3751367.
- ↑ Fournier-Viger, P.; Gomariz, A.; Gueniche, T. et al. (2014). "SPMF: A Java open-source pattern mining library". The Journal of Machine Learning Research 15 (1): 3389-3393. http://dl.acm.org/citation.cfm?id=2750353.
- ↑ Benites, F.; Sapozhnikova, E. (2014). "Evaluation of hierarchical interestingness measures for mining pairwise generalized association rules". IEEE Transactions on Knowledge and Data Engineering 26 (12): 3012-3025. doi:10.1109/TKDE.2014.2320722.
- ↑ Alaimo, S.; Pulvirenti, A.; Giugno, R.; Ferro, A.. "Drug-target interaction prediction through domain-tuned network-based inference". Bioinformatics 29 (16): 2004-8. doi:10.1093/bioinformatics/btt307. PMC PMC3722516. PMID 23720490. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3722516.
- ↑ "The R Project for Statistical Computing". The R Foundation. http://www.r-project.org/.
- ↑ Yaffe, M.B. (2002). "How do 14-3-3 proteins work? - Gatekeeper phosphorylation and the molecular anvil hypothesis". FEBS Letters 513 (1): 53–57. PMID 11911880.
- ↑ Zheng, N.; Schulman, B.A.; Song, L. et al. (2002). "Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex". Nature 416 (6882): 703–709. doi:10.1038/416703a. PMID 11961546.
- ↑ Ho, M.S.; Tsai, P.I.; Chien, C.T. (2006). "F-box proteins: The key to protein degradation". Journal of Biomedical Science 13 (2): 181-91. doi:10.1007/s11373-005-9058-2. PMID 16463014.
- ↑ Shannon, P.; Markiel, A.; Ozier, O. et al. (2003). "Cytoscape: A software environment for integrated models of biomolecular interaction networks". Genome Research 13 (11): 2498-504. doi:10.1101/gr.1239303. PMC PMC403769. PMID 14597658. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC403769.
- ↑ Liu, X.; Wu, J.; Gu, F.; Wang, J.; He, Z. (2015). "Discriminative pattern mining and its applications in bioinformatics". Briefings in Bioinformatics 16 (5): 884-900. doi:10.1093/bib/bbu042. PMID 25433466.
- ↑ Mehra, A.; Zahra, A.; Thompson, V. et al. (2013). "Mycobacterium tuberculosis type VII secreted effector EsxH targets host ESCRT to impair trafficking". PLoS Pathogens 9 (10): e1003734. doi:10.1371/journal.ppat.1003734. PMC PMC3814348. PMID 24204276. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3814348.
- ↑ Burian, J.; Yim, G.; Hsing, M. et al. (2013). "The mycobacterial antibiotic resistance determinant WhiB7 acts as a transcriptional activator by binding the primary sigma factor SigA (RpoV)". Nucleic Acids Research 41 (22): 10062-76. doi:10.1093/nar/gkt751. PMC PMC3905903. PMID 23990327. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3905903.
- ↑ Zheng, H.; Lu, L.; Wang, B. et al. (2008). "Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv". PLoS One 3 (6): e2375. doi:10.1371/journal.pone.0002375. PMC PMC2440308. PMID 18584054. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2440308.
- ↑ Maus, C.E.; Plikaytis, B.B.; Shinnick, T.M. (2005). "Molecular analysis of cross-resistance to capreomycin, kanamycin, amikacin, and viomycin in Mycobacterium tuberculosis". Antimicrobial Agents and Chemotherapy 49 (8): 3192-7. doi:10.1128/AAC.49.8.3192-3197.2005. PMC PMC2440308. PMID PMC1196259. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2440308.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. Some figures and tables were moved slightly up from their original position to correspond more closely to their reference in the text.