Difference between revisions of "Main Page/Featured article of the week/2016"
Shawndouglas (talk | contribs) (Created as needed.) |
Shawndouglas (talk | contribs) (Added last week's article of the week.) |
||
| Line 17: | Line 17: | ||
|<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: January 4–10:</h2> | |<br /><h2 style="font-size:105%; font-weight:bold; text-align:left; color:#000; padding:0.2em 0.4em; width:50%;">Featured article of the week: January 4–10:</h2> | ||
<div style="padding:0.4em 1em 0.3em 1em;"> | <div style="padding:0.4em 1em 0.3em 1em;"> | ||
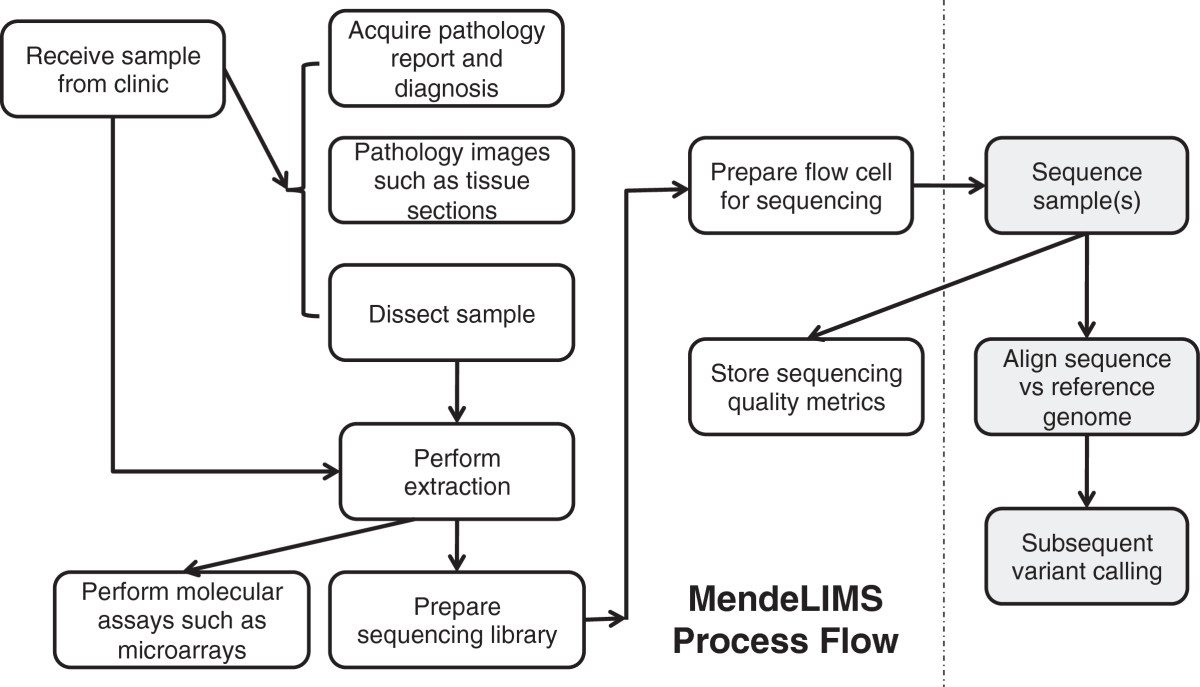
<div style="float: left; margin: 0.5em 0.9em 0.4em 0em;">[[File:Fig3 Grimes BMCBioinformatics2014 15.jpg|220px]]</div> | |||
'''"[[Journal:MendeLIMS: A web-based laboratory information management system for clinical genome sequencing|MendeLIMS: A web-based laboratory information management system for clinical genome sequencing]]"''' | |||
Large clinical genomics studies using next generation DNA sequencing require the ability to select and track samples from a large population of patients through many experimental steps. With the number of clinical genome sequencing studies increasing, it is critical to maintain adequate [[laboratory information management system]]s to manage the thousands of patient samples that are subject to this type of genetic analysis. | |||
To meet the needs of clinical population studies using genome sequencing, we developed a web-based laboratory information management system (LIMS) with a flexible configuration that is adaptable to continuously evolving experimental protocols of next generation DNA sequencing technologies. Our system is referred to as [[Stanford University School of Medicine#MendeLIMS|MendeLIMS]], is easily implemented with open source tools and is also highly configurable and extensible. MendeLIMS has been invaluable in the management of our clinical genome sequencing studies. ('''[[Journal:MendeLIMS: A web-based laboratory information management system for clinical genome sequencing|Full article...]]''')<br /> | |||
</div> | </div> | ||
|- | |- | ||
|<br /> | |||
|} | |} | ||
|} | |} | ||
<br /> | <br /> | ||
<div align="center">[[Main Page|— Return to the front page —]]</div> | <div align="center">[[Main Page|— Return to the front page —]]</div> | ||
Revision as of 19:56, 11 January 2016
|
|
If you're looking for the 2014 archive, it can be found here. The 2015 archive is here. |
Featured article of the week archive - 2016
Welcome to the LIMSwiki 2016 archive for the Featured Article of the Week.
|