|
If you're looking for the 2014 archive, it can be found here. The 2015 archive is here. |
Featured article of the week archive - 2016
Welcome to the LIMSwiki 2016 archive for the Featured Article of the Week.
Featured article of the week: January 18–24:
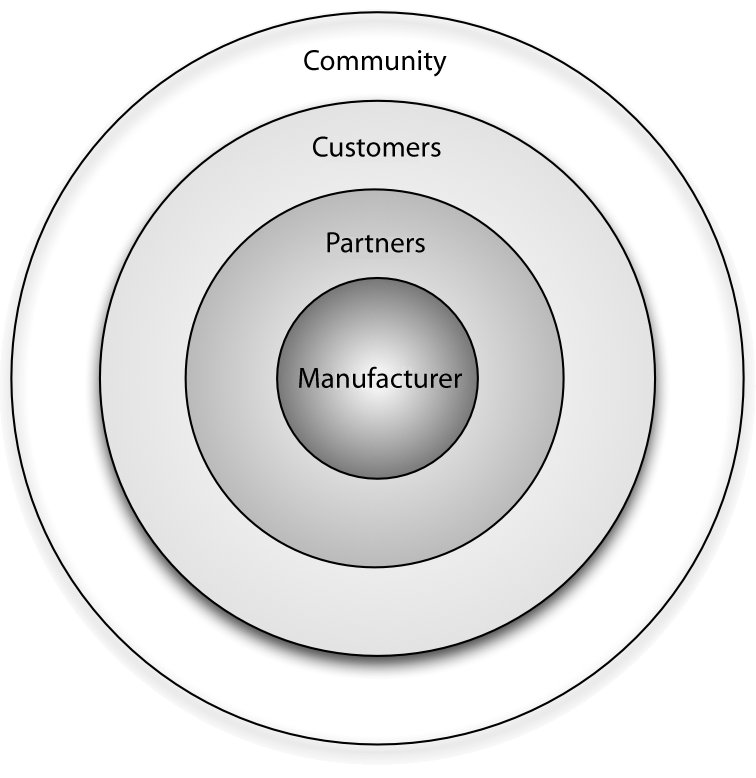
"Benefits of the community for partners of open source vendors"
Open source vendors can benefit from business ecosystems that form around their products. Partners of such vendors can utilize this ecosystem for their own business benefit by understanding the structure of the ecosystem, the key actors and their relationships, and the main levers of profitability. This article provides information on all of these aspects and identifies common business scenarios for partners of open source vendors. Armed with this information, partners can select a strategy that allows them to participate in the ecosystem while also maximizing their gains and driving adoption of their product or solution in the marketplace.
Every free/libre open source software (F/LOSS) vendor strives to create a business ecosystem around its software product. Doing this offers two primary advantages from a sales and marketing perspective: i) it increases the viability and longevity of the product in both commercial and communal spaces, and ii) it opens up new channels for communication and innovation. (Full article...)
Featured article of the week: January 11–17:
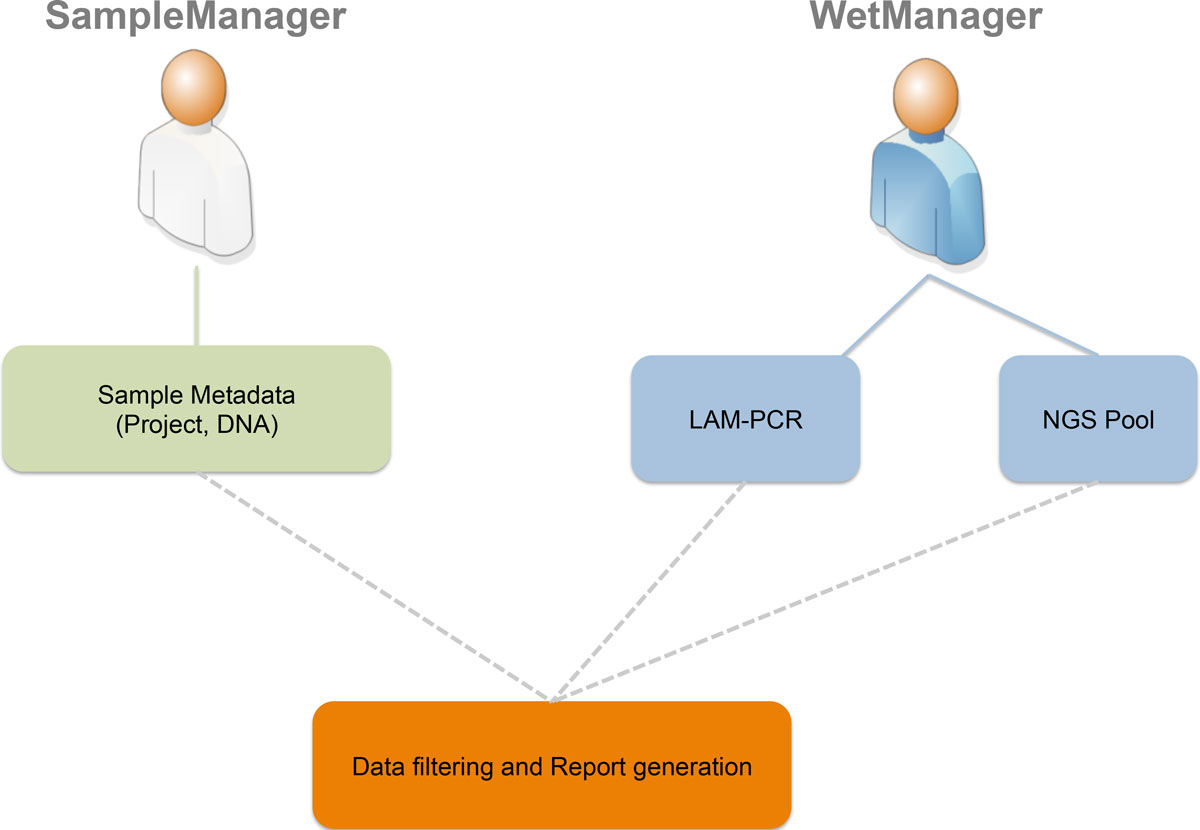
"adLIMS: A customized open source software that allows bridging clinical and basic molecular research studies"
Many biological laboratories that deal with genomic samples are facing the problem of sample tracking, both for pure laboratory management and for efficiency. Our laboratory exploits PCR techniques and Next Generation Sequencing (NGS) methods to perform high-throughput integration site monitoring in different clinical trials and scientific projects. Because of the huge amount of samples that we process every year, which result in hundreds of millions of sequencing reads, we need to standardize data management and tracking systems, building up a scalable and flexible structure with web-based interfaces, which are usually called Laboratory Information Management System (LIMS).
We extended and customized ADempiere ERP to fulfill LIMS requirements and we developed adLIMS. It has been validated by our end-users verifying functionalities and GUIs through test cases for PCRs samples and pre-sequencing data and it is currently in use in our laboratories. adLIMS implements authorization and authentication policies, allowing multiple users management and roles definition that enables specific permissions, operations and data views to each user. (Full article...)
|
Featured article of the week: January 4–10:
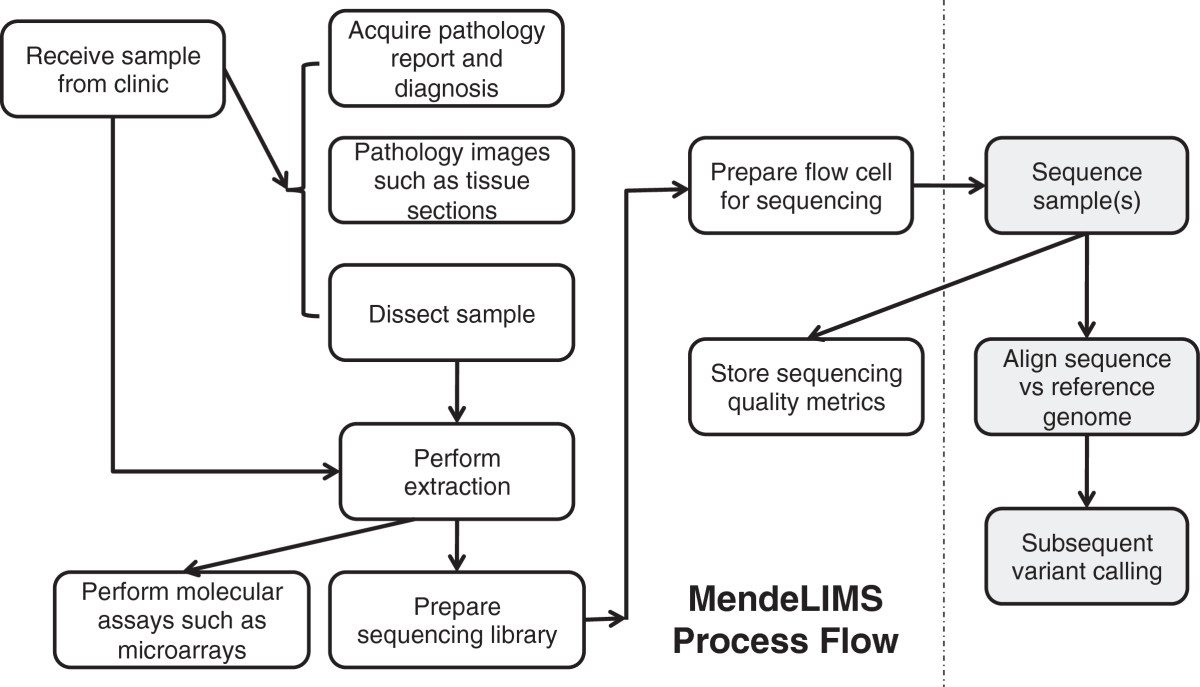
"MendeLIMS: A web-based laboratory information management system for clinical genome sequencing"
Large clinical genomics studies using next generation DNA sequencing require the ability to select and track samples from a large population of patients through many experimental steps. With the number of clinical genome sequencing studies increasing, it is critical to maintain adequate laboratory information management systems to manage the thousands of patient samples that are subject to this type of genetic analysis.
To meet the needs of clinical population studies using genome sequencing, we developed a web-based laboratory information management system (LIMS) with a flexible configuration that is adaptable to continuously evolving experimental protocols of next generation DNA sequencing technologies. Our system is referred to as MendeLIMS, is easily implemented with open source tools and is also highly configurable and extensible. MendeLIMS has been invaluable in the management of our clinical genome sequencing studies. (Full article...)
|
|