Featured article of the week: March 9–15:"What is the "source" of open-source hardware?"
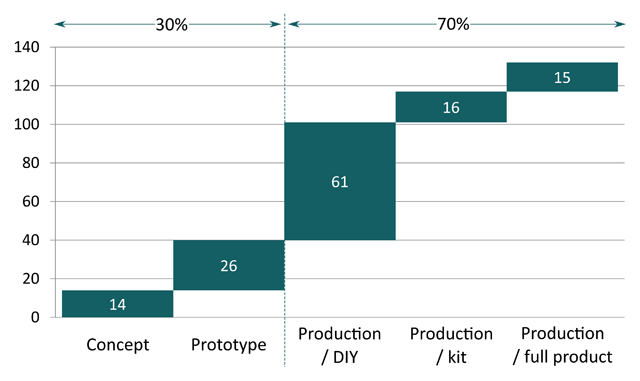
What “open source” means once applied to tangible products has been so far mostly addressed through the light of licensing. While this approach is suitable for software, it appears to be over-simplistic for complex hardware products. Whether such a product can be labelled as open-source is not only a question of licence but a question of documentation, i.e. what is the information that sufficiently describes it? Or in other words, what is the “source” of open-source hardware? To date there is no simple answer to this question, leaving large room for interpretation in the usage of the term. Based on analysis of public documentation of 132 products, this paper provides an overview of how practitioners tend to interpret the concept of open-source hardware. It specifically focuses on the recent evolution of the open-source movement outside the domain of electronics and DIY to that of non-electronic and complex open-source hardware products. The empirical results strongly indicate the existence of two main usages of open-source principles in the context of tangible products: publication of product-related documentation as a means to support community-based product development and to disseminate privately developed innovations. (Full article...)
Featured article of the week: March 2–8:
"From command-line bioinformatics to bioGUI"
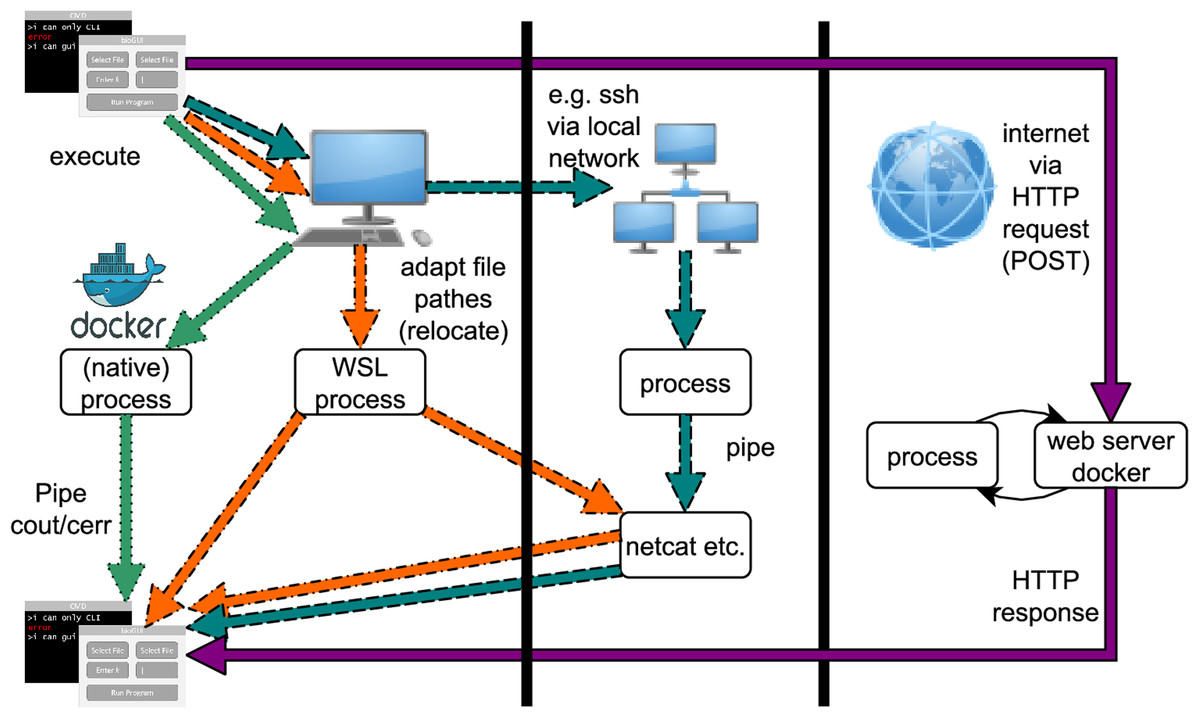
Bioinformatics is a highly interdisciplinary field providing informatics applications for scientists from many disciplines. Installing and starting applications on the command line (CL) is inconvenient and inefficient for many scientists. Nonetheless, most methods are implemented with a command-line interface only. Providing a graphical user interface (GUI) for bioinformatics applications is one step toward routinely making CL-only applications more readily available to scientists, yielding a positive step toward more effective interdisciplinary work. With our bioGUI framework, we address two main problems of using CL bioinformatics applications. First, many tools work on UNIX-based systems only, while many scientists use Microsoft Windows. Second, scientists refrain from using CL tools, which, despite their reservations, could well support them in their research. With bioGUI install modules and templates, installing and using CL tools is made possible for most scientists, even on Windows, due to bioGUI’s support for Windows Subsystem for Linux. In addition, bioGUI templates can easily be created, making the bioGUI framework highly rewarding for developers. From the bioGUI repository it is possible to download, install, and use bioinformatics tools with just a few clicks. (Full article...)
|
Featured article of the week: February 24–March 1:
"ChromaWizard: An open-source image analysis software for multicolor fluorescence in situ hybridization analysis"
Multicolor image analysis finds its applications in a broad range of biological studies. Specifically, multiplex fluorescence in situ hybridization (M‐FISH) for chromosome painting facilitates the analysis of individual chromosomes in complex metaphase spreads and is widely used to detect both numerical and structural aberrations. While this is well established for human and mouse karyotypes, for which species sophisticated software and analysis tools are available, other organisms and species are less well served. Commercially available software is proprietary and not easily adaptable to other karyotypes. Therefore, a publicly available open-source software that combines flexibility and customizable functionalities is needed. Here we present such a tool, called “ChromaWizard,” which is based on popular scientific image analysis libraries (OpenCV, scikit‐image, and NumPy). We demonstrate its functionality on the example of primary Chinese hamster (Cricetulus griseus) fibroblasts metaphase spreads and on Chinese hamster ovary cell lines, known for their large number of chromosomal rearrangements. (Full article...)
|
Featured article of the week: February 17–23:
"Haves and have nots must find a better way: The case for open scientific hardware"
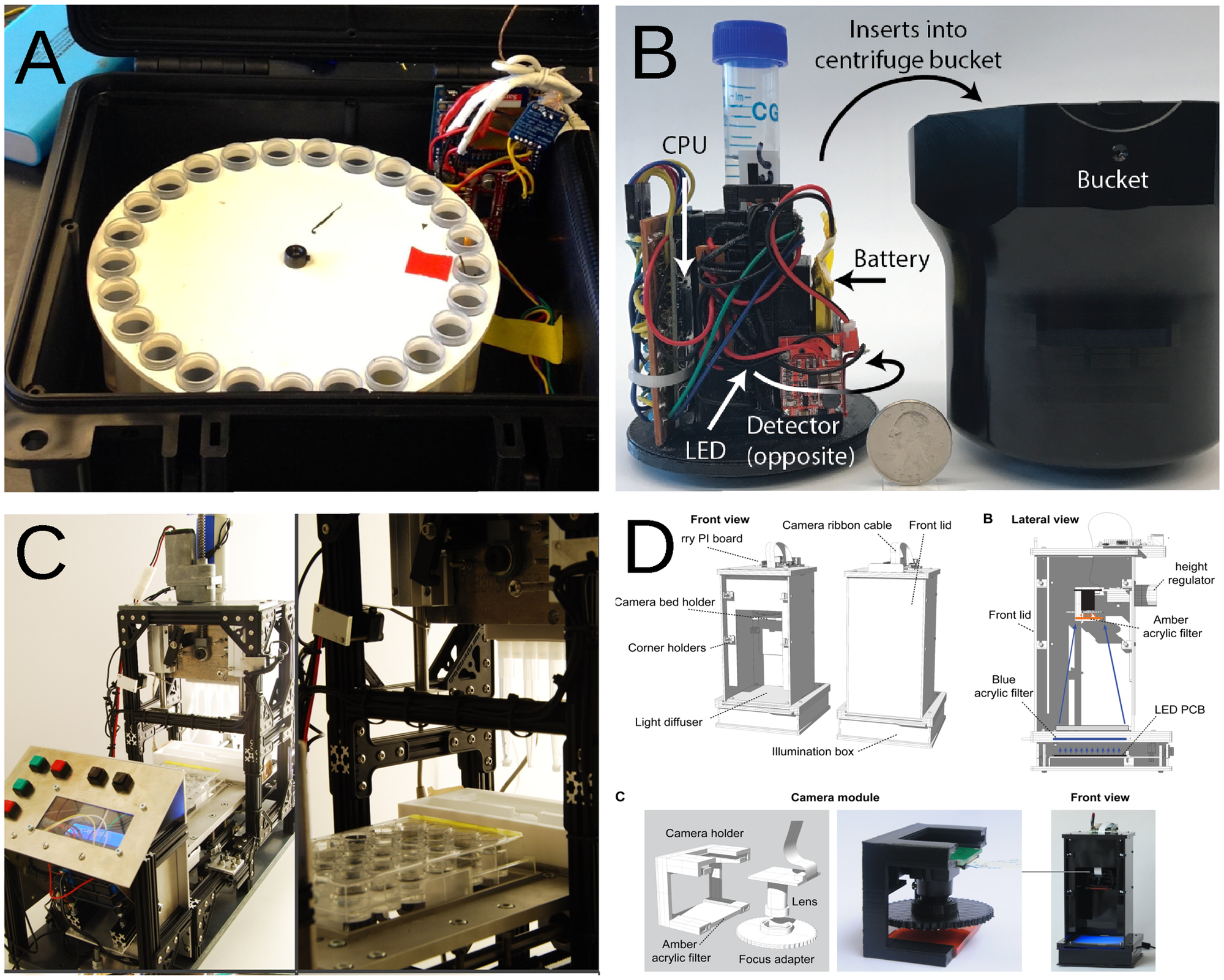
Many efforts are making science more open and accessible; they are mostly concentrated on issues that appear before and after experiments are performed: open access journals, open databases, and many other tools to increase reproducibility of science and access to information. However, these initiatives do not promote access to scientific equipment necessary for experiments. Mostly due to monetary constraints, equipment availability has always been uneven around the globe, affecting predominantly low-income countries and institutions. Here, a case is made for the use of free open-source hardware in research and education, including countries and institutions where funds were never the biggest problem. (Full article...)
|
Featured article of the week: February 03–16:
"CytoConverter: A web-based tool to convert karyotypes to genomic coordinates"
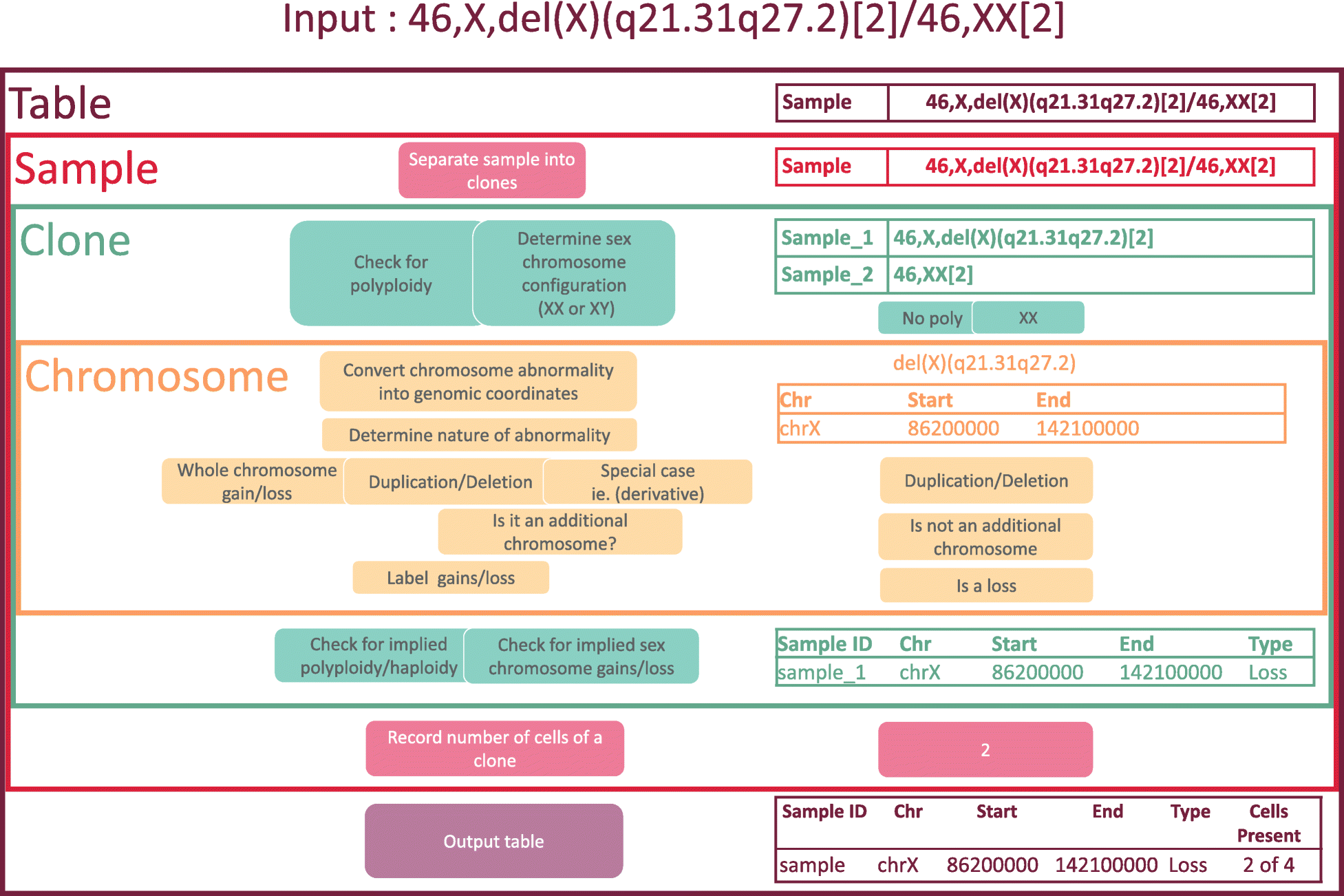
Cytogenetic nomenclature is used to describe chromosomal aberrations (or lack thereof) in a collection of cells, referred to as the cells’ karyotype. The nomenclature identifies locations on chromosomes using a system of cytogenetic bands, each with a unique name and region on a chromosome. Each band is microscopically visible after staining, and it encompasses a large portion of the chromosome. More modern analyses employ genomic coordinates, which precisely specify a chromosomal location according to its distance from the end of the chromosome. Currently, there is no tool to convert cytogenetic nomenclature into genomic coordinates. Since locations of genes and other genomic features are usually specified by genomic coordinates, a conversion tool will facilitate the identification of the features that are harbored in the regions of chromosomal gain and loss that are implied by a karyotype. (Full article...)
|
Featured article of the week: January 27–February 02:
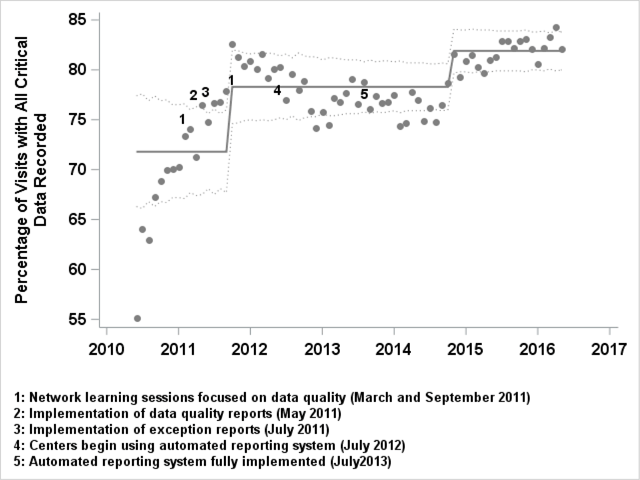
"Implementing a novel quality improvement-based approach to data quality monitoring and enhancement in a multipurpose clinical registry"
There is growing interest in the potential for clinical registries that can simultaneously support clinical care, quality improvement (QI), and research. This multi-purpose model is consistent with the Institute of Medicine’s (IOM’s) vision of a learning health system which “draws research closer to clinical practice by building knowledge development and application into each stage of the health care delivery process.” Gliklich et al. define a registry as “an organized system that uses observational study methods to collect uniform data (clinical and other) to evaluate specified outcomes for a population defined by a particular disease, condition, or exposure, and that serves one or more predetermined scientific, clinical, or policy purposes.” Most pediatric chronic illnesses meet the National Institutes of Health's (NIH) definition for rare disease, and as such, multi-center registries are especially important to study and improve care for children with chronic diseases. (Full article...)
|
Featured article of the week: January 20–26:
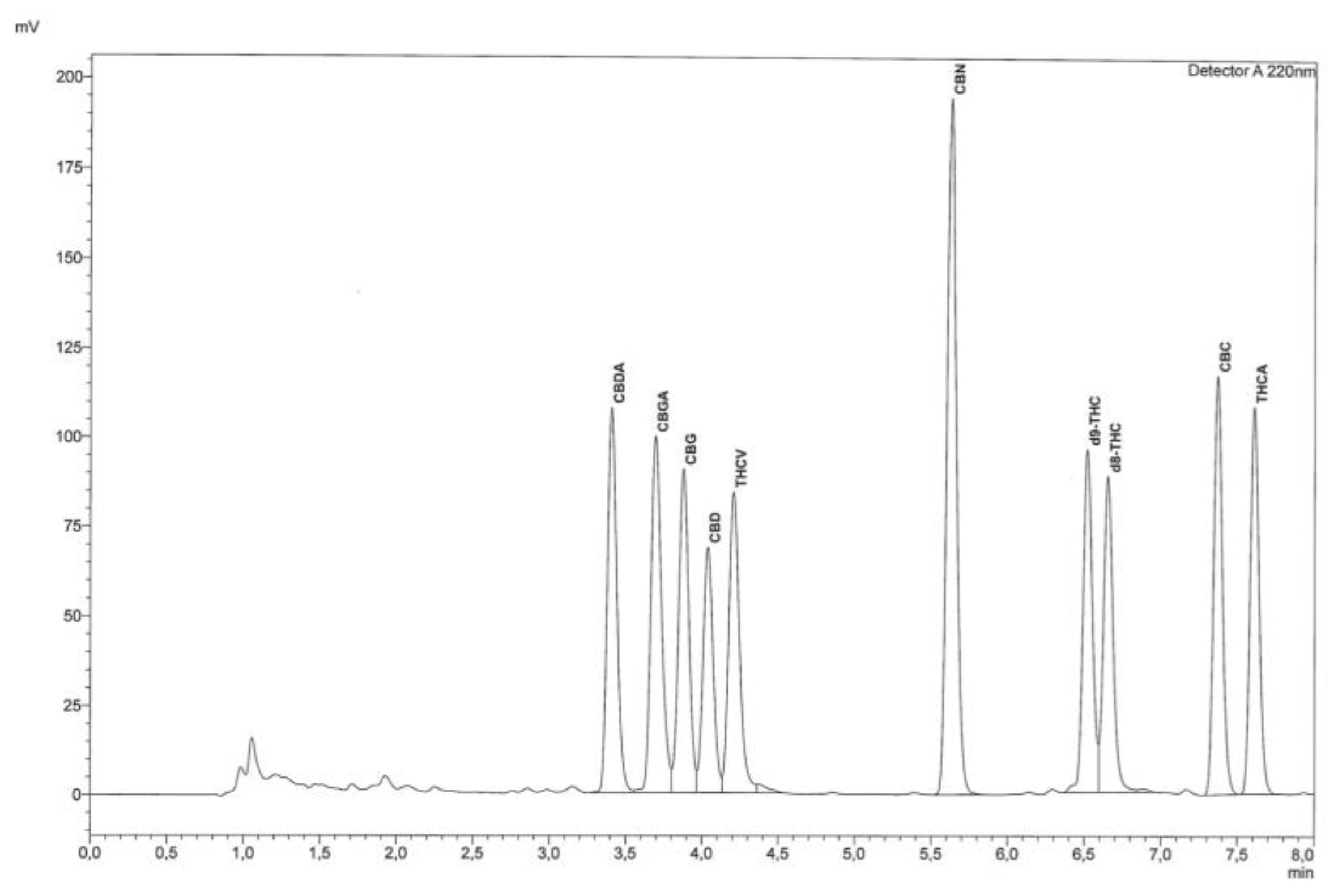
"Fast detection of 10 cannabinoids by RP-HPLC-UV method in Cannabis sativa L."
Cannabis has regained much attention as a result of updated legislation authorizing many different uses, and it can be classified on the basis of the content of Δ9-tetrahydrocannabinol (Δ9-THC), a psychotropic substance for which there are legal limitations in many countries. For this purpose, accurate qualitative and quantitative determination is essential. The relationship between THC and cannabidiol (CBD) is also significant, as the latter substance is endowed with many specific and non-psychoactive proprieties. For these reasons, it becomes increasingly important and urgent to utilize fast, easy, validated, and harmonized procedures for determination of cannabinoids. The procedure described herein allows rapid determination of 10 cannabinoids from the inflorescences of Cannabis sativa L. by extraction with organic solvents. Separation and subsequent detection are by reversed-phase high-performance liquid chromatography with ultraviolet detector (RP-HPLC-UV). (Full article...)
|
Featured article of the week: January 13–19:
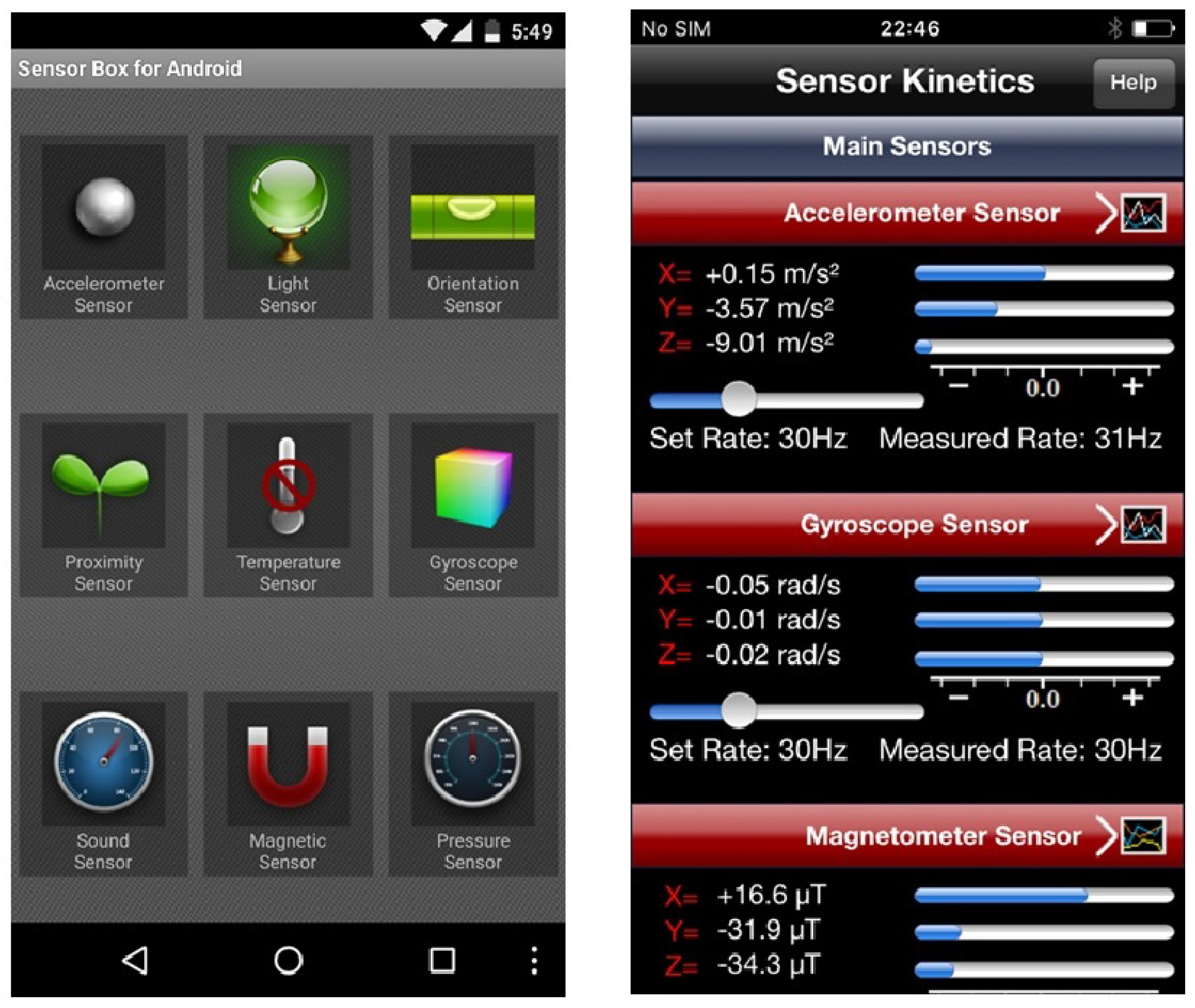
"What is this sensor and does this app need access to it?"
Mobile sensors have already proven to be helpful in different aspects of people’s everyday lives such as fitness, gaming, navigation, etc. However, illegitimate access to these sensors results in a malicious program running with an exploit path. While users are benefiting from richer and more personalized apps, the growing number of sensors introduces new security and privacy risks to end-users and makes the task of sensor management more complex. In this paper, we first discuss the issues around the security and privacy of mobile sensors. We investigate the available sensors on mainstream mobile devices and study the permission policies that Android, iOS and mobile web browsers offer for them. Second, we reflect on the results of two workshops that we organized on mobile sensor security. In these workshops, the participants were introduced to mobile sensors by working with sensor-enabled apps. We evaluated the risk levels perceived by the participants for these sensors after they understood the functionalities of these sensors. The results showed that knowing sensors by working with sensor-enabled apps would not immediately improve the users’ security inference of the actual risks of these sensors. However, other factors such as the prior general knowledge about these sensors and their risks had a strong impact on the users’ perception. (Full article...)
|
Featured article of the week: January 2–12:
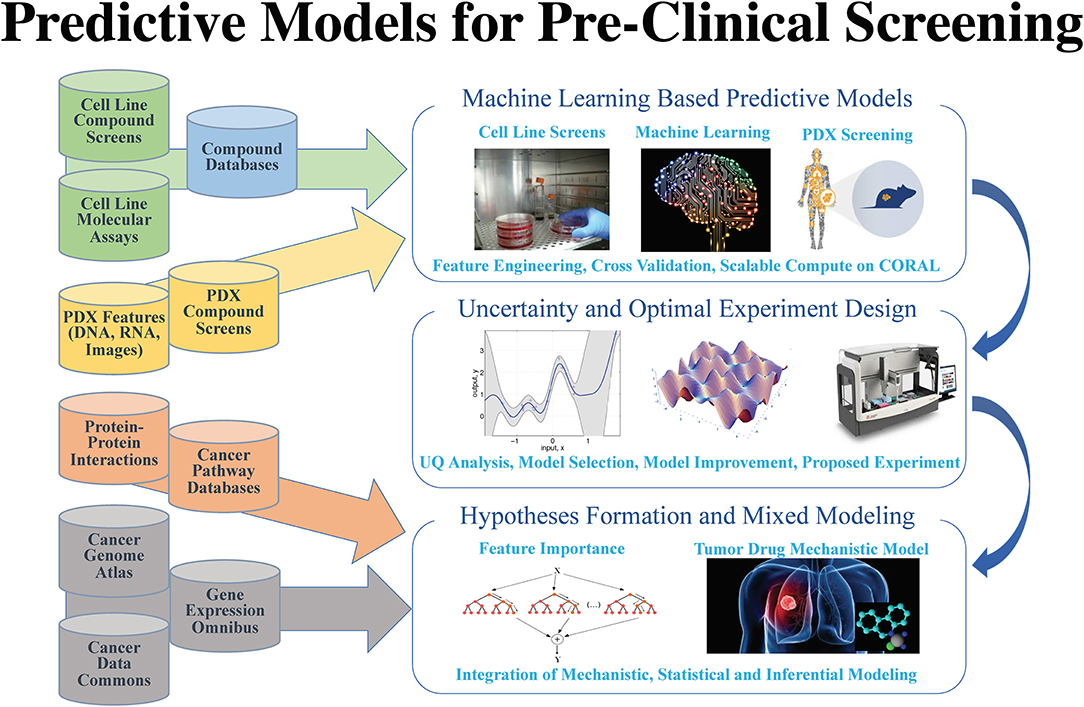
"AI meets exascale computing: Advancing cancer research with large-scale high-performance computing"
The application of data science in cancer research has been boosted by major advances in three primary areas: (1) data: diversity, amount, and availability of biomedical data; (2) advances in artificial intelligence (AI) and machine learning (ML) algorithms that enable learning from complex, large-scale data; and (3) advances in computer architectures allowing unprecedented acceleration of simulation and machine learning algorithms. These advances help build in silico ML models that can provide transformative insights from data, including molecular dynamics simulations, next-generation sequencing, omics, imaging, and unstructured clinical text documents. Unique challenges persist, however, in building ML models related to cancer, including: (1) access, sharing, labeling, and integration of multimodal and multi-institutional data across different cancer types; (2) developing AI models for cancer research capable of scaling on next-generation high-performance computers; and (3) assessing robustness and reliability in the AI models. (Full article...)
|
|