Difference between revisions of "User:Shawndouglas/sandbox/sublevel1"
Shawndouglas (talk | contribs) (Finished adding rest of content.) |
Shawndouglas (talk | contribs) |
||
| Line 3: | Line 3: | ||
| type = notice | | type = notice | ||
| style = width: 960px; | | style = width: 960px; | ||
| text = This is | | text = This is sublevel2 of my sandbox, where I play with features and test MediaWiki code. If you wish to leave a comment for me, please see [[User_talk:Shawndouglas|my discussion page]] instead.<p></p> | ||
}} | }} | ||
==Sandbox begins below== | ==Sandbox begins below== | ||
{{Infobox journal article | {{Infobox journal article | ||
|name = | |name = | ||
| Line 13: | Line 12: | ||
|alt = <!-- Alternative text for images --> | |alt = <!-- Alternative text for images --> | ||
|caption = | |caption = | ||
|title_full = | |title_full = A data quality strategy to enable FAIR, programmatic access across large,<br />diverse data collections for high performance data analysis | ||
|journal = '' | |journal = ''Informatics'' | ||
|authors = | |authors = Evans, Ben; Druken, Kelsey; Wang, Jingbo; Yang, Rui; Richards, Clare; Wyborn, Lesley | ||
|affiliations = | |affiliations = Australian National University | ||
|contact = Email: | |contact = Email: Jingbo dot Wang at anu dot edu dot au | ||
|editors = | |editors = Ge, Mouzhi; Dohnal, Vlastislav | ||
|pub_year = | |pub_year = 2017 | ||
|vol_iss = ''' | |vol_iss = '''4'''(4) | ||
|pages = | |pages = 45 | ||
|doi = [http://10. | |doi = [http://10.3390/informatics4040045 10.3390/informatics4040045] | ||
|issn = | |issn = 2227-9709 | ||
|license = [http://creativecommons.org/licenses/by/4.0/ Creative Commons Attribution 4.0 International] | |license = [http://creativecommons.org/licenses/by/4.0/ Creative Commons Attribution 4.0 International] | ||
|website = [ | |website = [http://www.mdpi.com/2227-9709/4/4/45/htm http://www.mdpi.com/2227-9709/4/4/45/htm] | ||
|download = [ | |download = [http://www.mdpi.com/2227-9709/4/4/45/pdf http://www.mdpi.com/2227-9709/4/4/45/pdf] (PDF) | ||
}} | }} | ||
{{ombox | {{ombox | ||
| Line 33: | Line 32: | ||
| text = This article should not be considered complete until this message box has been removed. This is a work in progress. | | text = This article should not be considered complete until this message box has been removed. This is a work in progress. | ||
}} | }} | ||
==Abstract== | |||
To ensure seamless, programmatic access to data for high-performance computing (HPC) and [[Data analysis|analysis]] across multiple research domains, it is vital to have a methodology for standardization of both data and services. At the Australian National Computational Infrastructure (NCI) we have developed a data quality strategy (DQS) that currently provides processes for: (1) consistency of data structures needed for a high-performance data (HPD) platform; (2) [[quality control]] (QC) through compliance with recognized community standards; (3) benchmarking cases of operational performance tests; and (4) [[quality assurance]] (QA) of data through demonstrated functionality and performance across common platforms, tools, and services. By implementing the NCI DQS, we have seen progressive improvement in the quality and usefulness of the datasets across different subject domains, and demonstrated the ease by which modern programmatic methods can be used to access the data, either ''in situ'' or via web services, and for uses ranging from traditional analysis methods through to emerging machine learning techniques. To help increase data re-usability by broader communities, particularly in high-performance environments, the DQS is also used to identify the need for any extensions to the relevant international standards for interoperability and/or programmatic access. | |||
'''Keywords''': data quality, quality control, quality assurance, benchmarks, performance, data management policy, netCDF, high-performance computing, HPC, fair data | |||
==Introduction== | ==Introduction== | ||
The National Computational Infrastructure (NCI) manages one of Australia’s largest and more diverse repositories (10+ petabytes) of research data collections spanning datasets from climate, coasts, oceans, and geophysics through to astronomy, [[bioinformatics]], and the social sciences.<ref name="WangLarge14">{{cite journal |title=Large-Scale Data Collection Metadata Management at the National Computation Infrastructure |journal=Proceedings from the American Geophysical Union, Fall Meeting 2014 |author=Wang, J.; Evans, B.J.K.; Bastrakova, I. et al. |pages=IN14B-07 |year=2014}}</ref> Within these domains, data can be of different types such as gridded, ungridded (i.e., line surveys, point clouds), and raster image types, as well as having diverse coordinate reference projections and resolutions. NCI has been following the Force 11 FAIR data principles to make data findable, accessible, interoperable, and reusable.<ref name="F11FAIR">{{cite web |url=https://www.force11.org/group/fairgroup/fairprinciples |title=The FAIR Data Principles |publisher=Force11 |accessdate=23 August 2017}}</ref> These principles provide guidelines for a research data repository to enable data-intensive science, and enable researchers to answer problems such as how to trust the scientific quality of data and determine if the data is usable by their software platform and tools. | |||
To ensure broader reuse of the data and enable transdisciplinary integration across multiple domains, as well as enabling programmatic access, a dataset must be usable and of value to a broad range of users from different communities.<ref name="EvansExtend16">{{cite journal |title=Extending the Common Framework for Earth Observation Data to other Disciplinary Data and Programmatic Access |journal=Proceedings from the American Geophysical Union, Fall General Assembly 2016 |author=Evans, B.J.K.; Wyborn, L.A.; Druken, K.A. et al. |pages=IN22A-05 |year=2016}}</ref> Therefore, a set of standards and "best practices" for ensuring the quality of scientific data products is a critical component in the life cycle of data management. We undertake both QC through compliance with recognized community standards (e.g., checking the header of the files to make sure it is compliant with community convention standard) and QA of data through demonstrated functionality and performance across common platforms, tools, and services (e.g., verifying the data to be functioning with designated software and libraries). | |||
The Earth Science Information Partners (ESIP) Information Quality Cluster (IQC) has been established for collecting such standards and best practices and then assisting data producers in their implementation, and users in their taking advantage of them.<ref name="RamapriyanEnsuring17">{{cite journal |title=Ensuring and Improving Information Quality for Earth Science Data and Products |journal=D-Lib Magazine |author=Ramapriyan, H.; Peng, G.; Moroni, D.; Shie, C.-L. |volume=23 |issue=7/8 |year=2017 |doi=10.1045/july2017-ramapriyan}}</ref> ESIP considers four different aspects of [[information]] quality in close relation to different stages of data products in their four-stage life cycle<ref name="RamapriyanEnsuring17" />: (1) define, develop, and validate; (2) produce, access, and deliver; (3) maintain, preserve, and disseminate; and (4) enable use, provide support, and service. | |||
Science teams or data producers are responsible for managing data quality during the first two stages, while data publishers are responsible for the latter two stages. As NCI is both a digital repository, which manages the storage and distribution of reference data for a range of users, as well as the provider of high-end compute and data analysis platforms, the data quality processes are focused on the latter two stages. A check on the scientific correctness is considered to be part of the first two stages and is not included in the definition of "data quality" that is described in this paper. | |||
==NCI's data quality strategy (DQS)== | |||
NCI developed a DQS to establish a level of assurance, and hence confidence, for our user community and key stakeholders as an integral part of service provision.<ref name="AtkinTotal05">{{cite book |chapter=Chapter 8: Service Specifications, Service Level Agreements and Performance |title=Total Facilities Management |author=Atkin, B.; Brooks, A. |publisher=Wiley |isbn=9781405127905}}</ref> It is also a step on the pathway to meet the technical requirements of a trusted digital repository, such as the CoreTrustSeal certification.<ref name="CTSData">{{cite web |url=https://www.coretrustseal.org/why-certification/requirements/ |title=Data Repositories Requirements |publisher=CoreTrustSeal |accessdate=24 October 2017}}</ref> As meeting these requirements involves the systematic application of agreed policies and procedures, our DQS provides a suite of guidelines, recommendations, and processes for: (1) consistency of data structures suitable for the underlying high-performance data (HPD) platform; (2) QC through compliance with recognized community standards; (3) benchmarking performance using operational test cases; and (4) QA through demonstrated functionality and benchmarking across common platforms, tools, and services. | |||
NCI’s DQS was developed iteratively through firstly a review of other approaches for management of data QC and data QA (e.g., Ramapriyan ''et al.''<ref name="RamapriyanEnsuring17" /> and Stall<ref name="StallAGU16">{{cite web |url=https://www.scidatacon.org/2016/sessions/100/ |title=AGU's Data Management Maturity Model |work=Auditing of Trustworthy Data Repositories |author=Stall, S.; Downs, R.R.; Kempler, S.J. |publisher=SciDataCon 2016 |date=2016}}</ref>) to establish the DQS methodology and secondly applying this to selected use cases at NCI which captured existing and emerging requirements, particularly the use cases that relate to HPC. | |||
Our approach is consistent with the American Geophysical Union (AGU) Data Management Maturity (DMM)SM model<ref name="StallAGU16" /><ref name="StallTheAmerican16">{{cite journal |title=The American Geophysical Union Data Management Maturity Program |journal=Proceedings from the eResearch Australasia Conference 2016 |author=Stall, S.; Hanson, B.; Wyborn, L. |pages=72 |year=2016 |url=https://eresearchau.files.wordpress.com/2016/03/eresau2016_paper_72.pdf}}</ref>, which was developed in partnership the Capability Maturity Model Integration (CMMI) Institute and adapted for their DMMSM<ref name="CMMIDataMan">{{cite web |url=https://cmmiinstitute.com/store/data-management-maturity-(dmm) |title=Data Management Maturity (DMM) |publisher=CMMI Institute LLC}}</ref> model for applications in the Earth and space sciences. The AGU DMMSM model aims to provide guidance on how to improve data quality and consistency and facilitate reuse in the data life cycle. It enables both producers of data and repositories that store data to ensure that datasets are "fit-for-purpose," repeatable, and trustworthy. The Data Quality Process Areas in the AGU DMMSM model define a collaborative approach for receiving, assessing, cleansing, and curating data to ensure "fitness" for intended use in the scientific community. | |||
After several iterations, the NCI DQS was established as part of the formal data publishing process and is applied throughout the cycle from submission of data to the NCI repository through to its final publication. The approach is also being adopted by the data producers who now engage with the process from the preparation stage, prior to ingestion onto the NCI data platform. Early consultation and feedback has greatly improved both the quality of the data as well as the timeliness for publication. To improve the efficiency further, one of our major data suppliers is including our DQS requirements in their data generation processes to ensure data quality is considered earlier in data production. | |||
The technical requirements and implementation of our DQS will be described as four major but related data components: structure, QC, benchmarking, and QA. | |||
===Data structure=== | |||
NCI's research data collections are particularly focused on enabling programmatic access, required by: (1) NCI core services such as the NCI supercomputer and NCI cloud-based capabilities; (2) community virtual [[Laboratory|laboratories]] and virtual research environments; (3) those that require remote access through established scientific standards-based protocols that use data services; and, (4) increasingly, by international data federations. To enable these different types of programmatic access, datasets must be registered in the central NCI catalogue<ref name="NCIDataPortal">{{cite web |url=https://geonetwork.nci.org.au/geonetwork/srv/eng/catalog.search#/home |title=NCI Data Portal |publisher=National Computational Infrastructure}}</ref>, which records their location for access both on the filesystems and via data services. | |||
This requires the data to be well-organized and compliant with uniform, professionally managed standards and consistent community conventions wherever possible. For example, the climate community Coupled Model Intercomparison Project (CMIP) experiments use the Data Reference Syntax (DRS)<ref name="TaylorCMIP12">{{cite web |url=https://pcmdi.llnl.gov/mips/cmip5/docs/cmip5_data_reference_syntax.pdf |format=PDF |title=CMIP5 Data Reference Syntax (DRS) and Controlled Vocabularies |author=Taylor, K.E.; Balaji, V.; Hankin, S. et al. |publisher=Program for Climate Model Diagnosis & Intercomparison |date=13 June 2012}}</ref>, whilst the National Aeronautics and Space Administration (NASA) recommends a specific name convention for Landsat satellite image products.<ref name="USGSLandsat">{{cite web |url=https://landsat.usgs.gov/what-are-naming-conventions-landsat-scene-identifiers |title=What are the naming conventions for Landsat scene identifiers? |publisher=U.S. Geological Survey |accessdate=23 August 2017}}</ref> The NCI data collection catalogue manages the details of each dataset through a uniform application of ISO 19115:2003<ref name="ISO19115">{{cite web |url=https://www.iso.org/standard/53798.html |title=ISO 19115-1:2014 Geographic information -- Metadata -- Part 1: Fundamentals |publisher=International Organization for Standardization |date=April 2014 |accessdate=25 May 2016}}</ref>, an international schema used for describing geographic information and services. Essentially, each catalogue entry points to the location of the data within the NCI data infrastructure. The catalogue entries also point to the services endpoints such as a standard data download point, data subsetting interface, as well as Open Geospatial Consortium (OGC) Web Mapping Service (WMS) and Web Coverage Services (WCS). NCI can publish data through several different servers, and as such the specific endpoint for each of these service capabilities is listed. | |||
NCI has developed a catalogue and directory policy, which provides guidelines for the organization of datasets within the concepts of data collections and data sub-collections and includes a comprehensive definition for each hierarchical layer. The definitions are: | |||
* A ''data collection'' is the highest in the hierarchy of data groupings at NCI. It is comprised of either an exclusive grouping of data subcollections, or it is a tiered structure with an exclusive grouping of lower tiered data collections, where the lowest tier data collection will only contain data subcollections. | |||
* A ''data subcollection'' is an exclusive grouping of datasets (i.e., belonging to only one subcollection) where the constituent datasets are tightly managed. It must have responsibilities within one organization with responsibility for the underlying management of its constituent datasets. A data subcollection constitutes a strong connection between the component datasets, and is organized coherently around a single scientific element (e.g., model, instrument). A subcollection must have compatible licenses such that constituent datasets do not need different access arrangements. | |||
* A ''dataset'' is a compilation of data that constitutes a programmable data unit that has been collected and organized using a self-contained process. For this purpose it must have a named data owner, a single license, one set of semantics, ontologies, vocabularies, and has a single data format and internal data convention. A dataset must include its version. | |||
* A ''dataset granule'' is used for some scientific domains that require a finer level of granularity (e.g., in satellite Earth Observation datasets). A granule refers to the smallest aggregation of data that can be independently described, inventoried, and retrieved as defined by NASA.<ref name="NASAGlossary">{{cite web |url=https://earthdata.nasa.gov/user-resources/glossary#ed-glossary-g |title=Granule |work=EarthData Glossary |accessdate=23 August 2017}}</ref> Dataset granules have their own metadata and support values associated with the additional attributes defined by parent datasets. | |||
In addition we use the term "data category" to identify common contents/themes across all levels of the hierarchy. | |||
* A ''data category'' allows a broad spectrum of options to encode relationships between data. A data category can be anything that weakly relates datasets, with the primary way of discovering the groupings within the data by key terms (e.g., keywords, attributes, vocabularies, ontologies). Datasets are not exclusive to a single category. | |||
=== | ====Organization of data within the data structure==== | ||
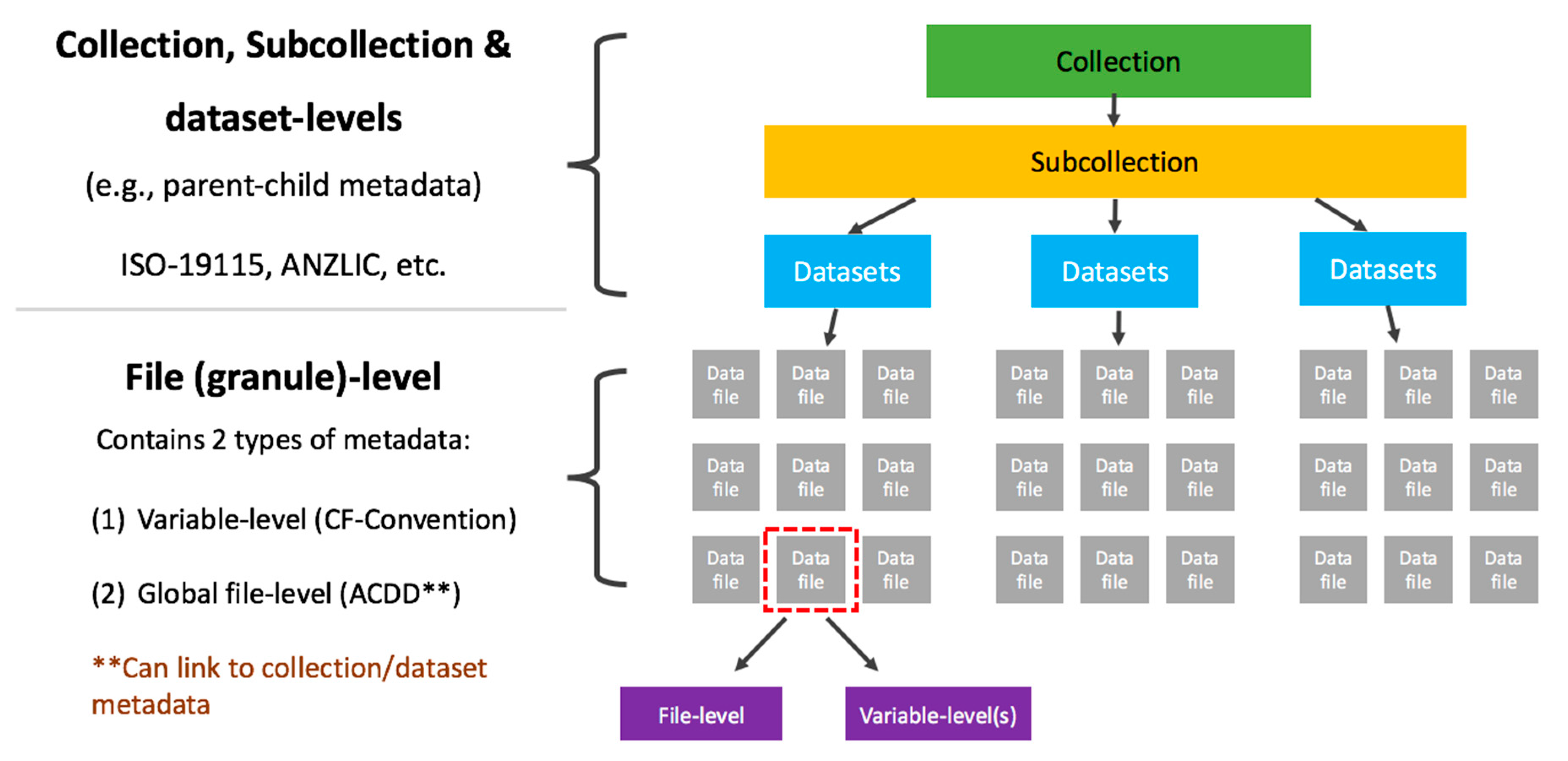
NCI has organized data collections according to this hierarchical structure on both filesystem and within our catalogue system. Figure 1 shows how these datasets are organized. Figure 2 provides an example of how the CMIP 5 data collection demonstrates the hierarchical directory structure. | |||
== | [[File:Fig1 Evans Informatics2017 4-4.png|700px]] | ||
{{clear}} | |||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| border="0" cellpadding="5" cellspacing="0" width="700px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 1.''' Illustration of the different levels of metadata and community standards used for each</blockquote> | |||
|- | |||
|} | |||
|} | |||
==References== | ==References== | ||
| Line 87: | Line 95: | ||
==Notes== | ==Notes== | ||
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. | This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. Several URL from the original were dead, and more current URLs were substituted. | ||
<!--Place all category tags here--> | <!--Place all category tags here--> | ||
[[Category:LIMSwiki journal articles (added in 2018)]] | [[Category:LIMSwiki journal articles (added in 2018)]] | ||
[[Category:LIMSwiki journal articles (all)]] | [[Category:LIMSwiki journal articles (all)]] | ||
[[Category:LIMSwiki journal articles on | [[Category:LIMSwiki journal articles on data quality]] | ||
[[Category:LIMSwiki journal articles on | [[Category:LIMSwiki journal articles on informatics]] | ||
Revision as of 18:23, 13 August 2018
|
|
This is sublevel2 of my sandbox, where I play with features and test MediaWiki code. If you wish to leave a comment for me, please see my discussion page instead. |
Sandbox begins below
| Full article title |
A data quality strategy to enable FAIR, programmatic access across large, diverse data collections for high performance data analysis |
|---|---|
| Journal | Informatics |
| Author(s) | Evans, Ben; Druken, Kelsey; Wang, Jingbo; Yang, Rui; Richards, Clare; Wyborn, Lesley |
| Author affiliation(s) | Australian National University |
| Primary contact | Email: Jingbo dot Wang at anu dot edu dot au |
| Editors | Ge, Mouzhi; Dohnal, Vlastislav |
| Year published | 2017 |
| Volume and issue | 4(4) |
| Page(s) | 45 |
| DOI | 10.3390/informatics4040045 |
| ISSN | 2227-9709 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | http://www.mdpi.com/2227-9709/4/4/45/htm |
| Download | http://www.mdpi.com/2227-9709/4/4/45/pdf (PDF) |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
To ensure seamless, programmatic access to data for high-performance computing (HPC) and analysis across multiple research domains, it is vital to have a methodology for standardization of both data and services. At the Australian National Computational Infrastructure (NCI) we have developed a data quality strategy (DQS) that currently provides processes for: (1) consistency of data structures needed for a high-performance data (HPD) platform; (2) quality control (QC) through compliance with recognized community standards; (3) benchmarking cases of operational performance tests; and (4) quality assurance (QA) of data through demonstrated functionality and performance across common platforms, tools, and services. By implementing the NCI DQS, we have seen progressive improvement in the quality and usefulness of the datasets across different subject domains, and demonstrated the ease by which modern programmatic methods can be used to access the data, either in situ or via web services, and for uses ranging from traditional analysis methods through to emerging machine learning techniques. To help increase data re-usability by broader communities, particularly in high-performance environments, the DQS is also used to identify the need for any extensions to the relevant international standards for interoperability and/or programmatic access.
Keywords: data quality, quality control, quality assurance, benchmarks, performance, data management policy, netCDF, high-performance computing, HPC, fair data
Introduction
The National Computational Infrastructure (NCI) manages one of Australia’s largest and more diverse repositories (10+ petabytes) of research data collections spanning datasets from climate, coasts, oceans, and geophysics through to astronomy, bioinformatics, and the social sciences.[1] Within these domains, data can be of different types such as gridded, ungridded (i.e., line surveys, point clouds), and raster image types, as well as having diverse coordinate reference projections and resolutions. NCI has been following the Force 11 FAIR data principles to make data findable, accessible, interoperable, and reusable.[2] These principles provide guidelines for a research data repository to enable data-intensive science, and enable researchers to answer problems such as how to trust the scientific quality of data and determine if the data is usable by their software platform and tools.
To ensure broader reuse of the data and enable transdisciplinary integration across multiple domains, as well as enabling programmatic access, a dataset must be usable and of value to a broad range of users from different communities.[3] Therefore, a set of standards and "best practices" for ensuring the quality of scientific data products is a critical component in the life cycle of data management. We undertake both QC through compliance with recognized community standards (e.g., checking the header of the files to make sure it is compliant with community convention standard) and QA of data through demonstrated functionality and performance across common platforms, tools, and services (e.g., verifying the data to be functioning with designated software and libraries).
The Earth Science Information Partners (ESIP) Information Quality Cluster (IQC) has been established for collecting such standards and best practices and then assisting data producers in their implementation, and users in their taking advantage of them.[4] ESIP considers four different aspects of information quality in close relation to different stages of data products in their four-stage life cycle[4]: (1) define, develop, and validate; (2) produce, access, and deliver; (3) maintain, preserve, and disseminate; and (4) enable use, provide support, and service.
Science teams or data producers are responsible for managing data quality during the first two stages, while data publishers are responsible for the latter two stages. As NCI is both a digital repository, which manages the storage and distribution of reference data for a range of users, as well as the provider of high-end compute and data analysis platforms, the data quality processes are focused on the latter two stages. A check on the scientific correctness is considered to be part of the first two stages and is not included in the definition of "data quality" that is described in this paper.
NCI's data quality strategy (DQS)
NCI developed a DQS to establish a level of assurance, and hence confidence, for our user community and key stakeholders as an integral part of service provision.[5] It is also a step on the pathway to meet the technical requirements of a trusted digital repository, such as the CoreTrustSeal certification.[6] As meeting these requirements involves the systematic application of agreed policies and procedures, our DQS provides a suite of guidelines, recommendations, and processes for: (1) consistency of data structures suitable for the underlying high-performance data (HPD) platform; (2) QC through compliance with recognized community standards; (3) benchmarking performance using operational test cases; and (4) QA through demonstrated functionality and benchmarking across common platforms, tools, and services.
NCI’s DQS was developed iteratively through firstly a review of other approaches for management of data QC and data QA (e.g., Ramapriyan et al.[4] and Stall[7]) to establish the DQS methodology and secondly applying this to selected use cases at NCI which captured existing and emerging requirements, particularly the use cases that relate to HPC.
Our approach is consistent with the American Geophysical Union (AGU) Data Management Maturity (DMM)SM model[7][8], which was developed in partnership the Capability Maturity Model Integration (CMMI) Institute and adapted for their DMMSM[9] model for applications in the Earth and space sciences. The AGU DMMSM model aims to provide guidance on how to improve data quality and consistency and facilitate reuse in the data life cycle. It enables both producers of data and repositories that store data to ensure that datasets are "fit-for-purpose," repeatable, and trustworthy. The Data Quality Process Areas in the AGU DMMSM model define a collaborative approach for receiving, assessing, cleansing, and curating data to ensure "fitness" for intended use in the scientific community.
After several iterations, the NCI DQS was established as part of the formal data publishing process and is applied throughout the cycle from submission of data to the NCI repository through to its final publication. The approach is also being adopted by the data producers who now engage with the process from the preparation stage, prior to ingestion onto the NCI data platform. Early consultation and feedback has greatly improved both the quality of the data as well as the timeliness for publication. To improve the efficiency further, one of our major data suppliers is including our DQS requirements in their data generation processes to ensure data quality is considered earlier in data production.
The technical requirements and implementation of our DQS will be described as four major but related data components: structure, QC, benchmarking, and QA.
Data structure
NCI's research data collections are particularly focused on enabling programmatic access, required by: (1) NCI core services such as the NCI supercomputer and NCI cloud-based capabilities; (2) community virtual laboratories and virtual research environments; (3) those that require remote access through established scientific standards-based protocols that use data services; and, (4) increasingly, by international data federations. To enable these different types of programmatic access, datasets must be registered in the central NCI catalogue[10], which records their location for access both on the filesystems and via data services.
This requires the data to be well-organized and compliant with uniform, professionally managed standards and consistent community conventions wherever possible. For example, the climate community Coupled Model Intercomparison Project (CMIP) experiments use the Data Reference Syntax (DRS)[11], whilst the National Aeronautics and Space Administration (NASA) recommends a specific name convention for Landsat satellite image products.[12] The NCI data collection catalogue manages the details of each dataset through a uniform application of ISO 19115:2003[13], an international schema used for describing geographic information and services. Essentially, each catalogue entry points to the location of the data within the NCI data infrastructure. The catalogue entries also point to the services endpoints such as a standard data download point, data subsetting interface, as well as Open Geospatial Consortium (OGC) Web Mapping Service (WMS) and Web Coverage Services (WCS). NCI can publish data through several different servers, and as such the specific endpoint for each of these service capabilities is listed.
NCI has developed a catalogue and directory policy, which provides guidelines for the organization of datasets within the concepts of data collections and data sub-collections and includes a comprehensive definition for each hierarchical layer. The definitions are:
- A data collection is the highest in the hierarchy of data groupings at NCI. It is comprised of either an exclusive grouping of data subcollections, or it is a tiered structure with an exclusive grouping of lower tiered data collections, where the lowest tier data collection will only contain data subcollections.
- A data subcollection is an exclusive grouping of datasets (i.e., belonging to only one subcollection) where the constituent datasets are tightly managed. It must have responsibilities within one organization with responsibility for the underlying management of its constituent datasets. A data subcollection constitutes a strong connection between the component datasets, and is organized coherently around a single scientific element (e.g., model, instrument). A subcollection must have compatible licenses such that constituent datasets do not need different access arrangements.
- A dataset is a compilation of data that constitutes a programmable data unit that has been collected and organized using a self-contained process. For this purpose it must have a named data owner, a single license, one set of semantics, ontologies, vocabularies, and has a single data format and internal data convention. A dataset must include its version.
- A dataset granule is used for some scientific domains that require a finer level of granularity (e.g., in satellite Earth Observation datasets). A granule refers to the smallest aggregation of data that can be independently described, inventoried, and retrieved as defined by NASA.[14] Dataset granules have their own metadata and support values associated with the additional attributes defined by parent datasets.
In addition we use the term "data category" to identify common contents/themes across all levels of the hierarchy.
- A data category allows a broad spectrum of options to encode relationships between data. A data category can be anything that weakly relates datasets, with the primary way of discovering the groupings within the data by key terms (e.g., keywords, attributes, vocabularies, ontologies). Datasets are not exclusive to a single category.
Organization of data within the data structure
NCI has organized data collections according to this hierarchical structure on both filesystem and within our catalogue system. Figure 1 shows how these datasets are organized. Figure 2 provides an example of how the CMIP 5 data collection demonstrates the hierarchical directory structure.
|
References
- ↑ Wang, J.; Evans, B.J.K.; Bastrakova, I. et al. (2014). "Large-Scale Data Collection Metadata Management at the National Computation Infrastructure". Proceedings from the American Geophysical Union, Fall Meeting 2014: IN14B-07.
- ↑ "The FAIR Data Principles". Force11. https://www.force11.org/group/fairgroup/fairprinciples. Retrieved 23 August 2017.
- ↑ Evans, B.J.K.; Wyborn, L.A.; Druken, K.A. et al. (2016). "Extending the Common Framework for Earth Observation Data to other Disciplinary Data and Programmatic Access". Proceedings from the American Geophysical Union, Fall General Assembly 2016: IN22A-05.
- ↑ 4.0 4.1 4.2 Ramapriyan, H.; Peng, G.; Moroni, D.; Shie, C.-L. (2017). "Ensuring and Improving Information Quality for Earth Science Data and Products". D-Lib Magazine 23 (7/8). doi:10.1045/july2017-ramapriyan.
- ↑ Atkin, B.; Brooks, A.. "Chapter 8: Service Specifications, Service Level Agreements and Performance". Total Facilities Management. Wiley. ISBN 9781405127905.
- ↑ "Data Repositories Requirements". CoreTrustSeal. https://www.coretrustseal.org/why-certification/requirements/. Retrieved 24 October 2017.
- ↑ 7.0 7.1 Stall, S.; Downs, R.R.; Kempler, S.J. (2016). "AGU's Data Management Maturity Model". Auditing of Trustworthy Data Repositories. SciDataCon 2016. https://www.scidatacon.org/2016/sessions/100/.
- ↑ Stall, S.; Hanson, B.; Wyborn, L. (2016). "The American Geophysical Union Data Management Maturity Program". Proceedings from the eResearch Australasia Conference 2016: 72. https://eresearchau.files.wordpress.com/2016/03/eresau2016_paper_72.pdf.
- ↑ "Data Management Maturity (DMM)". CMMI Institute LLC. https://cmmiinstitute.com/store/data-management-maturity-(dmm).
- ↑ "NCI Data Portal". National Computational Infrastructure. https://geonetwork.nci.org.au/geonetwork/srv/eng/catalog.search#/home.
- ↑ Taylor, K.E.; Balaji, V.; Hankin, S. et al. (13 June 2012). "CMIP5 Data Reference Syntax (DRS) and Controlled Vocabularies" (PDF). Program for Climate Model Diagnosis & Intercomparison. https://pcmdi.llnl.gov/mips/cmip5/docs/cmip5_data_reference_syntax.pdf.
- ↑ "What are the naming conventions for Landsat scene identifiers?". U.S. Geological Survey. https://landsat.usgs.gov/what-are-naming-conventions-landsat-scene-identifiers. Retrieved 23 August 2017.
- ↑ "ISO 19115-1:2014 Geographic information -- Metadata -- Part 1: Fundamentals". International Organization for Standardization. April 2014. https://www.iso.org/standard/53798.html. Retrieved 25 May 2016.
- ↑ "Granule". EarthData Glossary. https://earthdata.nasa.gov/user-resources/glossary#ed-glossary-g. Retrieved 23 August 2017.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. Several URL from the original were dead, and more current URLs were substituted.