Journal:SCIFIO: An extensible framework to support scientific image formats
| Full article title | SCIFIO: An extensible framework to support scientific image formats |
|---|---|
| Journal | BMC Bioinformatics |
| Author(s) | Hiner, Mark C.; Rueden, Curtis T.; Eliceiri, Kevin W. |
| Author affiliation(s) | University of Wisconsin at Madison, Morgridge Institute for Research |
| Primary contact | Email: eliceiri at wisc dot edu |
| Year published | 2016 |
| Volume and issue | 17 |
| Page(s) | 521 |
| DOI | 10.1186/s12859-016-1383-0 |
| ISSN | 1471-2105 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | http://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-016-1383-0 |
| Download | http://bmcbioinformatics.biomedcentral.com/track/pdf/10.1186/s12859-016-1383-0 (PDF) |
Abstract
Background: No gold standard exists in the world of scientific image acquisition; a proliferation of instruments each with its own proprietary data format has made out-of-the-box sharing of that data nearly impossible. In the field of light microscopy, the Bio-Formats library was designed to translate such proprietary data formats to a common, open-source schema, enabling sharing and reproduction of scientific results. While Bio-Formats has proved successful for microscopy images, the greater scientific community was lacking a domain-independent framework for format translation.
Results: SCIFIO (SCientific Image Format Input and Output) is presented as a freely available, open-source library unifying the mechanisms of reading and writing image data. The core of SCIFIO is its modular definition of formats, the design of which clearly outlines the components of image I/O to encourage extensibility, facilitated by the dynamic discovery of the SciJava plugin framework. SCIFIO is structured to support coexistence of multiple domain-specific open exchange formats, such as Bio-Formats’ OME-TIFF, within a unified environment.
Conclusions: SCIFIO is a freely available software library developed to standardize the process of reading and writing scientific image formats.
Keywords: SCIFIO, image analysis, open-source, Bio-Formats, ImageJ
Background
Image formats are defined by the logical layout of metadata and pixel information across one or more data sources. Proprietary file formats (PFFs) are created when an imaging instrument, such as a microscope, records such data in a structure that is not publicly described. PFFs are especially problematic in scientific domains, as each company or even instrument brings the potential for a new file format, possibly requiring licensed software to decode, or the file format changing in structure without notice or recourse. The scientific method necessitates that data can be analyzed by others to verify and reproduce results; when said data is stored in a proprietary format, by definition, it cannot be freely shared and inspected.
In response to the proliferation of PFFs in the fields of life science, the Open Microscopy Environment (OME) consortium developed the Bio-Formats library to standardize the reading of microscopy data.[1] Bio-Formats provides an application programming interface (API) for reading and writing images, backed by a comprehensive collection of extensions to decode format-specific information and translate it into an open specification called the OME data model.[2] A translated image can then be written as OME-TIFF, an “open-exchange format” which combines the universal readability of the TIFF standard with an XML schema representing the OME data model (OME-XML). These OME-TIFF images can be freely shared, with pixel data accessible via standard libraries such as libtiff[3], and the complete metadata parseable by any standards-compliant XML reader. In this way, the Bio-Formats project greatly mitigates the PFF problem in microscopy.
Bio-Formats has become an essential tool for scientists worldwide; however, its metadata model specifically targets 5-dimensional images in microscopy and related life sciences disciplines. PFFs from other scientific domains — e.g., medical imaging, astronomy, industrial x-rays, materials science and geoscience — each have their own unique considerations with respect to the dimensionality and metadata of their images; as such, it would be infeasible for a single “one-size-fits-all” metadata model to fully address the needs of scientific imaging as a whole. With this conclusion in mind, we have developed the SCIFIO (SCientific Image Format Input and Output) library, generalizing the success of Bio-Formats to create a domain-independent image I/O framework enabling seamless and extensible translation between image metadata models. The goal of SCIFIO is to provide the architecture that will equally facilitate: 1) the conversion of additional formats into supported open-exchange formats such as OME-TIFF and 2) the integration of additional scientific open-exchange formats such as Digital Imaging and Communications in Medicine (DICOM)[4], Flexible Image Transport System (FITS)[5] and NetCDF[6] into a common image I/O framework.
Implementation
SCIFIO is implemented as a plugin suite for the SciJava plugin framework. Its core is written under the permissive BSD license to maximize freedom of inclusion in both open- and closed-source applications. The SciJava framework collects Plugins in an application Context which are typically accessed via Services. As such, SCIFIO defines a collection of Plugins and Services facilitating image I/O. Developers will typically start with the SCIFIO class itself: a Gateway to the SciJava Context providing convenient access methods for functional components of the SCIFIO framework.
The SciJava framework sorts Plugins by “type,” representing the role of a given Plugin. Extensibility and flexibility is achieved by providing a public Service API which organizes and delegates to available Plugins of each type. Thus, SCIFIO development is primarily concerned with adding new Plugin implementations to achieve a desired result. The following sections describe the key Plugin types in SCIFIO, and the behavior they control.
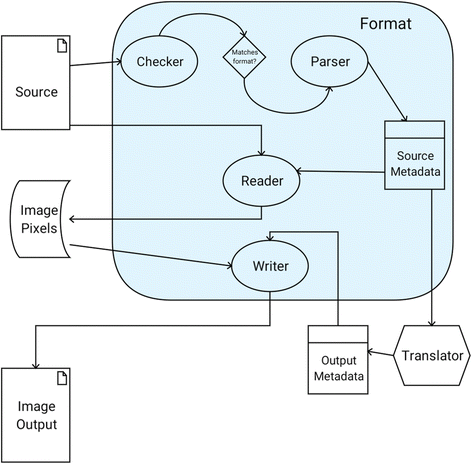
First and foremost is the Format. Formats are a collection of interface-driven components (Fig. 1) defining the steps for decoding an image source to its metadata and pixel values. In SCIFIO, the ImageJ Common data model is used to describe pixels; this data model is built on ImgLib2[7] due to its type and algorithmic flexibility, ensuring images opened with SCIFIO are universally recognized within the ImageJ ecosystem.[8] A Format must always include a Metadata component defining its unique fields and structures, such as acquisition instrument details, dimensional axis types, or detector emission wavelengths. Each Metadata implementation must also be able to express itself as a standard format-independent ImageMetadata object, establishing a common baseline for use within the framework.
|
The Checker component contains the logic for matching a given Format with a potential image source, while the Parser component performs the actual creation of Metadata from that source. The Reader and Writer components use Metadata to read or write pixel data, respectively. Given the goal of freely shareable image data, Writers are optional components and should not be implemented for proprietary formats.
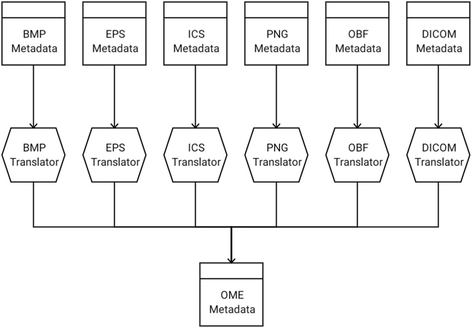
A second essential Plugin type is the Translator, which encodes logic for conversion from one Metadata type to another. Translators enable the standardization of proprietary formats to common Metadata structures such as OME, and hence play a key role in converting images between Formats. Translators are typically created to accompany Writers, ensuring Format-specific metadata is properly populated. Additionally, the Translator framework enables the integration of new open-exchange formats via Translator-only libraries, converting supported Metadata types to the new standard. An example of this model can be seen in the SCIFIO-OME-XML component (Fig. 2).
|
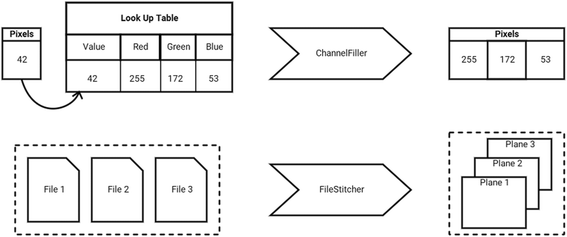
While Formats and Translators add new behavior to the base framework, SCIFIO also has Plugin types to control existing behavior. For example, Filter plugins provide a Format-agnostic mechanism for modifying Reader behavior. Filters create an ordered chain of delegation, each operating on the data of its parent, and can be individually toggled ‘on’ or ‘off’ on a per-Reader basis. Sample Filter stacking behavior is illustrated in a ChannelFiller for converting “indexed color” pixels to RGB values and a FileStitcher for unifying multiple files on disk to form one dataset (Fig. 3).
|
With all SciJava Plugins, a numeric priority value attached to each class creates an implicit relative ordering for operations — e.g., order of Checker querying, Translator querying, or Filter application. Priorities are automatically considered when using the SCIFIO Services: from the FormatService polling Checker components to the TranslatorService finding the correct Translator for a given request, priorities allow querying the most specific solutions first, before moving to more general options. These pieces together provide a robust and flexible library for reading and writing image data.
Results and discussion
As the fundamental goal of SCIFIO is to establish an extensible framework for image support, the SciJava framework is a logical choice for implementation. SciJava provides extensible solutions to common software problems, which implicitly benefit SCIFIO. A core example is the extensible script language framework (http://imagej.net/Scripting) which effectively allows SCIFIO to be used from any number of programming languages without requiring language-specific considerations in SCIFIO itself.
ImageJ[9] presents the flagship use case for SCIFIO, allowing an established community to vet and refine the library. Although users do not directly interact with SCIFIO API, all image I/O operations in ImageJ ultimately rely on SCIFIO. As developers contribute new Format plugins for image types relevant to their work, any application using SCIFIO can immediately benefit from the new plugin. Looking beyond ImageJ, projects like KNIME Image Processing (KNIP), built on the KNIME Analytics Platform[10], have already adopted SCIFIO for their image I/O mechanism. This sort of code sharing leads to a form of mutualistic collaboration: a new Format plugin developed for KNIP will automatically work in ImageJ, with the converse true as well. Equally importantly, both ImageJ and KNIP can implicitly operate on image data produced by the other program, laying the foundation for algorithmic interoperability.
Collaborations like this would not be possible with a focused library like Bio-Formats. KNIME is a platform for extensible workflows, thus its handling of image data demands flexibility beyond the fixed 5D microscopy schema of OME. Additionally, Bio-Formats’ mechanism of format extension requires either modification of a text-based configuration file to define format priority, which can lead to conflicts if multiple libraries provide differing versions of this file, or runtime modification by API calls, which may not be reproducible without a central mechanism controlling these calls. Conversely, the dynamic discovery of the SciJava plugin framework allows SCIFIO developers to provide their Formats completely independently — e.g., on an ImageJ, KNIME or Eclipse update site, while SCIFIO’s backing by the ImageJ Common data model ensures adaptation to any future requirements in imaging dimensionality and data types.
Bio-Formats readers and writers and SCIFIO Format components define similar high level logic, but in Bio-Formats several I/O steps are conflated in a single monolithic interface with many protected methods as potential extension points. SCIFIO encapsulates each I/O step into its own dedicated component, to minimize the effort required in format development. Whether a format is added to Bio-Formats or SCIFIO libraries; the SCIFIO-BF-Compat and SCIFIO-OME-XML components offer bidirectional compatibility between SCIFIO and Bio-Formats.
Bio-Formats has demonstrated the feasibility of standardizing a broad field of PFFs into a common open-exchange format. SCIFIO provides a natural generalization of thinking, allowing extension to new domains, through the integration of their Metadata standards and open-exchange formats via Translators, and clear paths for contributing to existing domains by encapsulating the logic of Format components. Given the added immediate power of the Bio-Formats integration layers, we see the SCIFIO framework as a potential unifying solution to PFFs in scientific image data.
Conclusions
SCIFIO is an open-source library generalizing the successful structure of Bio-Formats to create a domain-independent framework for the reading, writing, and translation of images. The extensible design of SCIFIO facilitates community contribution, the establishment of domain-specific metadata standards, and integration into a unified system capable of adapting to the demands of scientific imaging analysis.
Abbreviations
API: Application program interface
I/O: Input and/or output
KNIME: Konstanz Information Miner
KNIP: KNIME Image Processing
OME: Open Microscopy Environment
PFF: Proprietary file formats
SCIFIO: SCientific Image Format Input and Output
Declarations
Acknowledgements
Many people have contributed to the development of SCIFIO on both technical and leadership levels. In particular, the authors gratefully thank and acknowledge the efforts of (in alphabetical order): Ellen T. Arena, Anne Carpenter, Christian Dietz, Gabriel Einsdorf, Melissa Linkert, Josh Moore, Tobias Pietzsch, Stephan Preibisch, Stephan Saalfeld, Jason Swedlow, and Pavel Tomancak. We also thank the entire ImageJ community, especially those who contributed patch submissions, use cases, feature requests and bug reports.
Funding
Research reported in this publication was supported by ACI Division of Advanced Cyberinfrastructure of the National Science Foundation under award number 1148362 and additional internal funding from the Laboratory for Optical and Computational Instrumentation.
Availability of data and materials
Project name: SCIFIO
Project home page: http://scif.io/
Archived version: 0.28.2 http://maven.imagej.net/service/local/repositories/releases/content/io/scif/scifio/0.28.2/scifio-0.28.2.jar
Source code: https://github.com/scifio/scifio
Operating system(s): Platform-independent
Programming language: Java
Other requirements: Java 1.8 or higher runtime, io.scif:scifio-jai-imageio, net.imagej:imagej-common, net.imglib2:imglib2, org.scijava:scijava-common, org.mapdb:mapdb
License: BSD
Authors’ contributions
MCH was the lead implementer of the software. CTR architected the underlying SciJava foundation and guided SCIFIO development. As the primary principal investigator of SCIFIO, KWE directed and advised on all aspects of the project including development directions and priorities. All authors contributed to, read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Maven artifacts
SCIFIO can be added as a dependency to any project capable of consuming Maven dependencies. As SCIFIO is a project in the SciJava domain, we recommend using dependency management from the latest pom-scijava release (http://maven.imagej.net/index.html#nexus-search;gav~org.scijava~pom-scijava). The following are example sections for adding a SCIFIO dependency to a pom.xml:
References
- ↑ Linkert, M.; Rueden, C.T.; Allan, C. et al. (2010). "Metadata matters: Access to image data in the real world". Journal of Cell Biology 189 (5): 777–82. doi:10.1083/jcb.201004104. PMID 20513764.
- ↑ Goldberg, I.G.; Allan, C.; Burel, J.M. et al. (2005). "The Open Microscopy Environment (OME) Data Model and XML file: Open tools for informatics and quantitative analysis in biological imaging". Genome Biology 6 (5): R47. doi:10.1186/gb-2005-6-5-r47. PMID 15892875.
- ↑ Warmerdam, F.; Kiseley, A.; Welles, M.; Kelly, D.. "LibTIFF - TIFF Library and Utilities". http://www.libtiff.org/. Retrieved 29 November 2016.
- ↑ Bidgood Jr., W.D.; Horii, S.C.; Prior, F.W.; Van Syckle, D.E. (1997). "Understanding and using DICOM, the data interchange standard for biomedical imaging". JAMIA 4 (3): 199–212. doi:10.1136/jamia.1997.0040199. PMID 9147339.
- ↑ Pence, W.D.; Chiappetti, L.; Page, C.G. et al. (2010). "Definition of the Flexible Image Transport System (FITS), version 3.0". Astronomy & Astrophysics 524 (December 2010): A42. doi:10.1051/0004-6361/201015362.
- ↑ Unidata. "Network Common Data Form (NetCDF)". University Corporation for Atmospheric Research. doi:10.5065/D6H70CW6. http://www.unidata.ucar.edu/software/netcdf/. Retrieved 29 November 2016.
- ↑ Pietzsch, T.; Preisbisch, S.; Tomancák, P. et al. (2012). "ImgLib2: Generic image processing in Java". Bioinformatics 28 (22): 3009–11. doi:10.1093/bioinformatics/bts543. PMID 22962343.
- ↑ Schindelin, J.; Rueden, C.T.; Hiner, M.C. et al. (2015). "The ImageJ ecosystem: An open platform for biomedical image analysis". Molecular Reproduction and Development 82 (7–8): 518-29. doi:10.1002/mrd.22489. PMID 26153368.
- ↑ Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. (2012). "NIH Image to ImageJ: 25 years of image analysis". Nature Methods 9 (7): 671–5. PMID 22930834.
- ↑ Berthold, M.R.; Cebron, N.; Dill, F.; Gabriel, T.R.; Kötter, T.; Meinl, T.; Ohl, P.; Sieb, C.; Thiel, K.; Wiswedel, B. (2008). "Chapter 38: KNIME: The Konstanz Information Miner". In Preisach, C.; Burkhardt, H.; Schmidt-Thieme, L.; Decker, R.. Data Analysis, Machine Learning and Applications. Springer Berlin Heidelberg. doi:10.1007/978-3-540-78246-9_38. ISBN 9783540782391.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added.