Difference between revisions of "Journal:A web application to support the coordination of reflexive, interpretative toxicology testing"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) (Saving and adding more.) |
||
| Line 43: | Line 43: | ||
Yang ''et al.'' [9] described that adding interpretative comments from a trained laboratory director to urine drug screening results can greatly impact both the clinician and patient. Clinicians may not understand the analytical method limitations, and any resulting misinterpretation of the results may lead to a patient's stigmatization or inappropriate termination from care. The authors further reported an increase in urine drug screen orders with interpretive comments, indicating that clinicians value the interpretation made by a trained director. Because accurate interpretation of results often requires the integration of documented clinical information, medication data, test results, pharmacokinetics, and test limitations and interferences, assigning a trained [[Pathology|pathologist]] or [[Clinical chemistry|clinical chemist]] who is familiar with interpreting clinical [[information]], as well as the biochemistry and testing methods to provide an interpretation of the results, is a valuable tool in ensuring the correct diagnosis for patients. | Yang ''et al.'' [9] described that adding interpretative comments from a trained laboratory director to urine drug screening results can greatly impact both the clinician and patient. Clinicians may not understand the analytical method limitations, and any resulting misinterpretation of the results may lead to a patient's stigmatization or inappropriate termination from care. The authors further reported an increase in urine drug screen orders with interpretive comments, indicating that clinicians value the interpretation made by a trained director. Because accurate interpretation of results often requires the integration of documented clinical information, medication data, test results, pharmacokinetics, and test limitations and interferences, assigning a trained [[Pathology|pathologist]] or [[Clinical chemistry|clinical chemist]] who is familiar with interpreting clinical [[information]], as well as the biochemistry and testing methods to provide an interpretation of the results, is a valuable tool in ensuring the correct diagnosis for patients. | ||
One challenge laboratories and laboratory directors face with UDT is that there may be results from several tests that inform whether additional testing is required and need to be integrated to provide an interpretation given the specific clinical context. Within the Department of Laboratory Medicine and Pathology at University of Washington Medicine (UW Medicine), we have formulated a urine drug testing ordering protocol and interpretation workflow. Clinicians may select from a menu of panels that are based on the assessed patient's risk of treatment non-compliance. Panels include an [[immunoassay]] drug screen and confirmation LC-MS/MS assays for opioids, amphetamines, benzodiazepines, and alcohol. For example, a low-risk patient panel consists of an immunoassay drug screen, and aberrant or unexpected results can lead the reviewing pathologist to order one or more of the confirmation tests. On top of the interpretation challenges, turnaround times (TAT) for the different tests are unevenly distributed. Most [[laboratory information system]]s (LISs) are not equipped to track, collect, and display the different results in an effective and informative manner to support this workflow. | One challenge laboratories and laboratory directors face with UDT is that there may be results from several tests that inform whether additional testing is required and need to be integrated to provide an interpretation given the specific clinical context. Within the Department of Laboratory Medicine and Pathology at University of Washington Medicine (UW Medicine), we have formulated a urine drug testing ordering protocol and interpretation [[workflow]]. Clinicians may select from a menu of panels that are based on the assessed patient's risk of treatment non-compliance. Panels include an [[immunoassay]] drug screen and confirmation LC-MS/MS assays for opioids, amphetamines, benzodiazepines, and alcohol. For example, a low-risk patient panel consists of an immunoassay drug screen, and aberrant or unexpected results can lead the reviewing pathologist to order one or more of the confirmation tests. On top of the interpretation challenges, turnaround times (TAT) for the different tests are unevenly distributed. Most [[laboratory information system]]s (LISs) are not equipped to track, collect, and display the different results in an effective and informative manner to support this workflow. | ||
The aim of this project was to design and implement a web-based software application that centralizes test results for pathologist review, performs the [[quality control]] (QC) calculations for the complex LC-MS/MS analysis of opioids and their metabolites, functions as a user input form for pathologist interpretation entry, and auto-files results into the LIS. | The aim of this project was to design and implement a web-based software application that centralizes test results for pathologist review, performs the [[quality control]] (QC) calculations for the complex LC-MS/MS analysis of opioids and their metabolites, functions as a user input form for pathologist interpretation entry, and auto-files results into the LIS. | ||
==Methods== | ==Methods== | ||
The initial step before designing the application was to understand the entire workflow, then gather requirements and specifications from stakeholders. | |||
===Pre-application testing and workflows=== | |||
At UW Medicine, for each patient specimen undergoing the opiate confirmation assay, two extractions are prepared, an undiluted and a diluted 10-fold urine sample. To help improve the complex LC-MS/MS [[data analysis]] of opioids and opioid metabolites, a command-line software application was developed. [10] The software application known as “SMACK” was shown to improve the data analysis process by automating a QC algorithm and calculations to improve consistency of analysis as well as reduce the amount of time medical laboratory scientists (MLS) spent reviewing the data. | |||
The algorithm was part of an intricate workflow that involved several steps, spreadsheets, and individuals. The goal of the spreadsheets was to collect and consolidate the relevant patient information so that the reviewing pathologist had the necessary information to generate an interpretation for the overall case. The information with the relevant patient data was spread across several spreadsheets, the creation of which relied on laboratory staff and pathologists copying and pasting various fields from one spreadsheet to another. Pathologists and laboratory staff communicated the completion of steps via email while using a naming convention for the relevant files. | |||
===Requirements=== | |||
We set to gather information on how clinicians ordered the different drug monitoring tests and how the information arrived at the laboratory. We investigated the LIS data workflow that was in place and how this information could feed the application, including assay results and patient demographics. Additionally, we worked with the laboratory to understand how test results were produced and how this information should be included in the application. Lastly, we investigated what the requirements were for progressing from one step of the workflow to the next along with the data associated with the progression. In addition to the workflow in the laboratory, we needed to understand what data elements are needed for each step of the workflow. With this information, we would create a map of the workflow to visualize and better understand the workflow. We gathered necessary components from stakeholders, pathologists, directors, pathologists in training, and laboratory staff by interviewing stakeholders and observing workflows. During this process, we gathered user interface requirements from stakeholders to shape functionality of the optimized web application. | |||
===Technical requirements=== | |||
The previously described QC software SMACK had been implemented as a command-line application in [[Python (programming language)|Python]], with no dependencies outside of the standard library. [10] Input data is provided as an [[Extensible Markup Language|XML]] format file exported from [[Waters Corporation|Waters]]' TargetLynx software. The outputs of the software are two CSV-formatted reports, one containing assay results and the other a detailed report of the QC calculations. Appendix A, Supplemental Fig. 1 displays the flow chart defining the QC algorithm, which has been simplified since its initial implementation. For this project, we needed to incorporate the software into the application to perform the same QC calculations on the opiate assay raw data and then display the output to the user in a meaningful manner. Moreover, our group has a defined technical software stack for developing and deploying applications in Python. For these reasons, writing the application in Python was the most efficient choice. An additional requirement was to automatically file results into the LIS (from [[CliniSys, Inc.]]). To achieve this requirement, we had to work with Data Innovations' Instrument Manager software. | |||
===Performance measurement and user feedback=== | |||
Finally, to assess the performance, we surveyed stakeholders and documented the rough amount of time that is required in each step of the workflow. Input was solicited from several laboratory staff members rotating through the area regarding how much time was spent on each step before and after the implementation of the application. We also surveyed the pathologists in training (chemistry fellows and rotating residents), known as trainees. Finally, we extracted data from the LIS to capture the number of occurrences in which a testing order had a correction or modification after the initial data entry so we could compare correction rates prior to and after implementation. | |||
==Results== | |||
===Laboratory workflows prior to implementation=== | |||
Providers order UDTs for the patient based on an assessment of the patient's risk for compliance. Table 1 displays the patient risk levels, the intended population, and the tests associated with each risk panel. | |||
{| class="wikitable" border="1" cellpadding="5" cellspacing="0" width="60%" | |||
|- | |||
| colspan="3" style="background-color:white; padding-left:10px; padding-right:10px;" |'''Table 1.''' Urine drug testing ordering strategy based on risk. Providers assess risk, review clinical patient care data, and order tests as described. | |||
|- | |||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |Risk | |||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |Intended population | |||
! style="background-color:#dddddd; padding-left:10px; padding-right:10px;" |Tests | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Low | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Morphine equivalent dose <90 mg/day; Stable on chronic opioid therapy or buprenorphine therapy | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Drug screen immunoassay | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Medium to high | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Morphine equivalent dose >=90 mg/day; Aberrant behavior (including early prescriptions, lost prescriptions, outbursts in office) | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Drug screen immunoassay; Enzymatic assay for alcohol; Opiate confirmation LCMS | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |High | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Same as medium to high; added concern for clonazepam or lorazepam | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |Drug screen immunoassay; Enzymatic assay for alcohol; Opiate confirmation LCMS; Benzodiazepine confirmation LCMS | |||
|- | |||
|} | |||
For each panel, providers have the option to enter any drug the patient is expected to be taking which may influence the test results. Each test in the panel is conducted separately and the results are entered into the LIS. Once all the tests have been completed and results entered, the pathologist may begin generating an interpretation. | |||
When pathologists are reviewing the results, they consider the patient's history in the [[electronic health record]]s (EHRs) for information that may influence the test results such as medications not specified by the ordering provider. These notes would be entered in dedicated fields in the spreadsheet. For training purposes, a pathologist trainee will often review the results first, then review their work with a director who may discuss the case, edit the preliminary interpretation, then make the final approval. Upon review of cases that initially only received the immunoassay drug screen, a probable outcome is for the reviewing pathologist to deem additional confirmation testing necessary. Therefore, the workflow is the summation of two separate parts, cases which receive opiate confirmation testing versus those that do not. The consideration to separate the workflow into two parts was partly because many orders require additional reflexive orders which influences how quickly the final interpretations will be released. For this reason, it was best to group cases by risk level to provide an interpretation as quickly as possible. Laboratory staff worked with departmental software engineers to create and deliver spreadsheets for each workflow which contain the patient's demographics (medical record number, age, sex, name, and order number) along with the results of the tests. | |||
The opiate assay is performed on a Waters Xevo TQ-MS tandem mass spectrometer with an Aquity UPLC system. Data acquisition is controlled by Waters MassLynx and chromatography review is completed using Waters TargetLynx software. When the data acquisition is complete, the technologist completes a cursory review of the auto-integration using TargetLynx. Once the preliminary review is complete, an XML file with the raw data is created. The XML file is the input of the application that performs QC calculations from the LC-MS/MS raw data. The output was two detailed tabular reports; the first was the calculated results after the algorithm was applied and another with a detailed report of the QC calculations. These reports would be reviewed in Microsoft Excel by two laboratory staff members to address any values that did not meet QC standards before entering the results into the LIS. Once the results have been reviewed, staff would send an email to the reviewing pathologists that reports were ready for their review. The pathologist would then combine the calculated results with the spreadsheet containing other test results and patient demographics via a copy-paste method. During the pathologist review, they may identify aberrant results stemming from interfering compounds or possible adulterations based on the data. They would then create an email thread with the laboratory to communicate and resolve the issue. | |||
One possible outcome for those cases where patient risk levels are associated with low to medium risk, a reviewing pathologist orders a confirmatory test. Pathologists would type the test codes for the orders they wished to place in the spreadsheet and the reviewing MLS would place the orders in the LIS. Therefore, a third spreadsheet was created to capture the results for the cases that received additional tests. To transfer the notes captured during the initial review, a Microsoft Excel macro was created which would parse the spreadsheets and bring the pathologist's saved work from the initial spreadsheet into the final spreadsheet. | |||
These spreadsheets were created and delivered via a dedicated website where staff and faculty were able to download the spreadsheets from any computer in the laboratory. After the spreadsheet was initially downloaded and edited, the file was stored on a shared drive accessible through a protected network that was accessible to the laboratory and pathologists. | |||
A naming convention for these files was created to help indicate the stage in the workflow. | |||
Once the reviewing pathologist had completed their review, they would send an email to staff so that they could enter the interpretation result into the LIS manually. If the technologist identified any issues with the interpretation, they would email the pathologist to edit/review the interpretation. | |||
===Technical requirements=== | |||
The application was implemented in Python using the Flask web framework<ref name="FlaskProj">{{cite web |url=https://flask.palletsprojects.com/en/2.3.x/ |title=Flask |publisher=Pallets |date=2010}}</ref> and other open-source libraries. Appendix A, Supplemental Table 1 lists the required libraries used for the application. | |||
The software was developed on an Apple Macintosh running Mac OS 10.15 and is used in production on Amazon Web Services (AWS) server running Ubuntu (18.04 LTS). The data from the LIS is gathered using Cache Object Script that collects information every 30 minutes from 6 a.m. to 6 p.m. and is delivered to a secure cloud storage resource (S3 bucket) on AWS. Once the results are finalized, the application sends a tab-delimited file to a transfer Linux (4.15.0) server which is picked up by Data Innovations instrument manager that sends the data to the LIS. | |||
An important component of the software development and deployment process was the use of the Git version control software. Git is a free, open-source, distributed version control software that can be used to store “snapshots” of source code files in which modifications are captured to the code repository. Each change is attributed to a specific author and can be associated with a comment describing the intent of the change. A unique tag identifies each change, and the application captures the tag in the software version number and records this identifier to every data output. In this manner, results of each analysis can be traced to the exact version of the software used to collect and generate those results. Additionally, [[GitLab]] is a Git-based fully integrated platform which allows users to create “issues” where enhancements, development, and other topics may be discussed collectively. This feature of GitLab can also attribute labels to each issue so that larger topics or concepts can be collectively viewed and labeled. | |||
The application is a web-based application that can be launched from any web browser. Access control is managed using our institutional identity provider with single sign-on (SSO) with two-factor authentication (2FA). For auto-resulting, once the results reach their terminal status (pathologists and laboratory staff have completed their collective review), a tab-separated-values (TSV) is sent to Data Innovations' Instrument Manager where it files the results to the LIS. | |||
===Requirements=== | |||
The data collected and workflows described serviced as the foundation of the application's design. The application needed to gather the necessary data from the LIS and display the information in a manner that is useful to the user. A system needed to be designed to progress a case through the necessary steps for the appropriate reviewer. Entry fields needed to be available for all members to be able to communicate problems, capture notes by the pathologists during their review, and for the final interpretation result. The application needed to ingest the XML file from TargetLynx of the opiate test, apply the SMACK algorithm, display both the QC calculations and results, and save all edits. Lastly, the application needed a system to communicate any additional confirmation tests the pathologist wanted to order. | |||
A new component within the laboratory workflow was the introduction of an [[Laboratory automation|automated liquid handler]] to prepare the urine samples for the opiate assay. The laboratory had been manually preparing the samples and entering the patient identifiers into the acquisition system, MassLynx. When creating the extraction method of the liquid handler, the decision was made to use the [[barcode]] on the specimen label as the sample identifier for the assay. To associate the result back to the patient, all specimen IDs also needed to be gathered. | |||
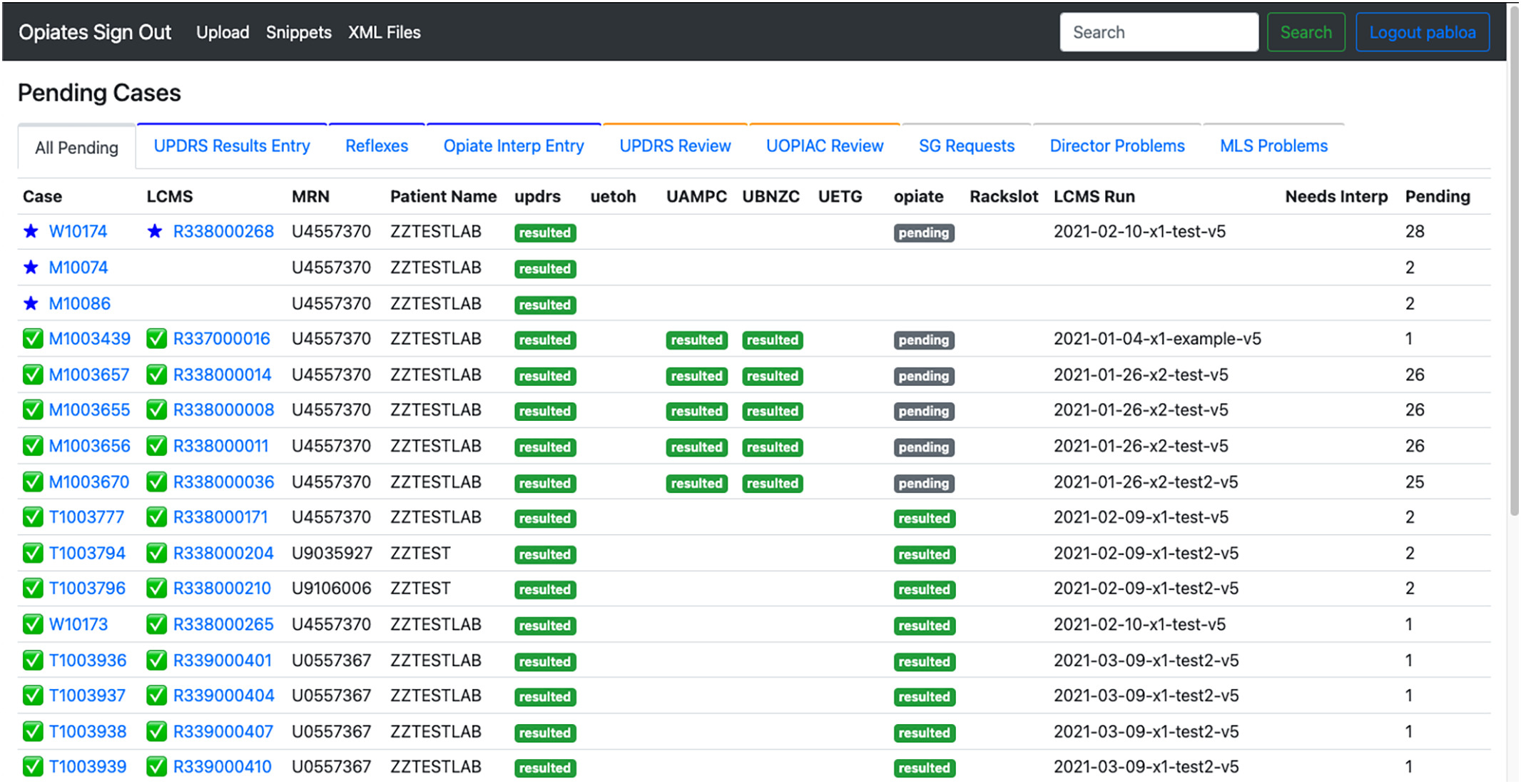
The decision to create a web-based application was to provide an easy-access entry point for the laboratory and pathologists. Also, a web-based application provided the flexibility needed to create a custom site that fit the needs of the workflow. Fig. 1 shows the home page of the application. | |||
[[File:Fig1 Pablo JofPathInfo2023 14.jpg|1000px]] | |||
{{clear}} | |||
{| | |||
| style="vertical-align:top;" | | |||
{| border="0" cellpadding="5" cellspacing="0" width="1000px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Figure 1.''' Home page of the opiate sign-out application. The landing page lists all pending samples in the application along with sample information such as case number, container ID (LCMS), medical record number, patient name, and status of each test.</blockquote> | |||
|- | |||
|} | |||
|} | |||
==References== | ==References== | ||
Revision as of 19:31, 13 June 2023
| Full article title | A web application to support the coordination of reflexive, interpretative toxicology testing |
|---|---|
| Journal | Journal of Pathology Informatics |
| Author(s) | Pablo, Abed; Laha, Thomas J.; Breit, Nathan; Hoffman, Noah G.; Hoofnagle, Andrew N.; Baird, Geoffrey S.; Mathias, Patrick C. |
| Author affiliation(s) | University of Washington School of Medicine |
| Primary contact | Email: pcm10 at uw dot edu |
| Year published | 2023 |
| Volume and issue | 14 |
| Article # | 100303 |
| DOI | 10.1016/j.jpi.2023.100303 |
| ISSN | 2153-3539 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.sciencedirect.com/science/article/pii/S2153353923001177 |
| Download | https://www.sciencedirect.com/science/article/pii/S2153353923001177/pdfft (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
Background: Reflexive laboratory testing workflows can improve the assessment of patients receiving pain medications chronically, but complex workflows requiring pathologist input and interpretation may not be well-supported by traditional laboratory information systems (LISs). In this work, we describe the development of a web application that improves the efficiency of pathologists and laboratory staff in delivering actionable toxicology results.
Method: Before designing the application, we set out to understand the entire workflow, including the laboratory workflow and pathologist review. Additionally, we gathered requirements and specifications from stakeholders. Finally, to assess the performance of the implementation of the application, we surveyed stakeholders and documented the approximate amount of time that is required in each step of the workflow.
Results: A web-based application was chosen for the ease of access for users. Relevant clinical data was routinely received and displayed in the application. The workflows in the laboratory and during the interpretation process served as the basis of the user interface (UI). With the addition of auto-filing software, the return on investment (ROI) was significant. The laboratory saved the equivalent of one full-time employee in time by automating file management and result entry.
Discussion: Implementation of a purpose-built application to support reflex and interpretation workflows in a clinical pathology practice has led to a significant improvement in laboratory efficiency. Custom- and purpose-built applications can help reduce staff burnout, reduce transcription errors, and allow staff to focus on more critical issues around quality.
Keywords: Python, laboratory workflows, custom web application, quality control, mass spectrometry
Background
Urine drug monitoring is an increasingly important tool for primary care physicians (PCPs) and pain specialists to monitor patients on chronic opioid therapy. [1] Routine and random monitoring for all patients on long-term opioid therapy is recommended prior to starting and throughout drug therapy. [2] There are various analytical methods and matrices available for patient monitoring. Urine drug testing (UDT) has become the “gold-standard” for detecting illicit drug use and monitoring ongoing therapy. Urine is non-invasive, readily available, and contains higher concentrations of drugs and metabolites compared with other matrices. Immunoassay testing, commonly referred to as urine drug screening, detects the presence of selected drugs and/or metabolites based on a detection threshold. Typically, the immunoassay functions as the initial evaluation for the potential detection of drugs, but because it lacks specificity for analytes of the same drug class, it can lead to false-positive and -negative results. Therefore, drug screens are often coupled with a confirmatory detection method for more specific testing. Chromatography is generally reserved for confirmatory or definitive testing following immunoassay screening. Historically, gas chromatography–mass spectrometry (GC–MS) was the standard for confirmatory testing. However, Liquid chromatography–mass spectrometry–tandem mass spectrometry (LC-MS/MS) has gained favor over GC-MS due to reduced complexity of sample preparation methods and the broader applicability of LC-MS/MS to different drug classes. [3, 4]
Many laboratories are equipped to support UDT. [1,3,4,8,9] A major challenge of staged drug testing (i.e., screening and confirmatory assays) is the review and interpretation of the large number of quantitative and qualitative results that is generated by screening and confirmatory methods. [5,6,7] Pharmacokinetic mechanisms dictate how quickly and how much of a drug and its metabolites appear in an individual's urine, which can be challenging for physicians who have limited understanding due to reviewing a limited number of these results. [5] An incorrect interpretation of the LC-MS/MS results could mistakenly suggest that a patient used a non-prescribed medication. Additionally, the potential for analytes to co-elute could affect the signal of the analytes, influencing the overall accuracy of the LC-MS/MS test. [4]
Yang et al. [9] described that adding interpretative comments from a trained laboratory director to urine drug screening results can greatly impact both the clinician and patient. Clinicians may not understand the analytical method limitations, and any resulting misinterpretation of the results may lead to a patient's stigmatization or inappropriate termination from care. The authors further reported an increase in urine drug screen orders with interpretive comments, indicating that clinicians value the interpretation made by a trained director. Because accurate interpretation of results often requires the integration of documented clinical information, medication data, test results, pharmacokinetics, and test limitations and interferences, assigning a trained pathologist or clinical chemist who is familiar with interpreting clinical information, as well as the biochemistry and testing methods to provide an interpretation of the results, is a valuable tool in ensuring the correct diagnosis for patients.
One challenge laboratories and laboratory directors face with UDT is that there may be results from several tests that inform whether additional testing is required and need to be integrated to provide an interpretation given the specific clinical context. Within the Department of Laboratory Medicine and Pathology at University of Washington Medicine (UW Medicine), we have formulated a urine drug testing ordering protocol and interpretation workflow. Clinicians may select from a menu of panels that are based on the assessed patient's risk of treatment non-compliance. Panels include an immunoassay drug screen and confirmation LC-MS/MS assays for opioids, amphetamines, benzodiazepines, and alcohol. For example, a low-risk patient panel consists of an immunoassay drug screen, and aberrant or unexpected results can lead the reviewing pathologist to order one or more of the confirmation tests. On top of the interpretation challenges, turnaround times (TAT) for the different tests are unevenly distributed. Most laboratory information systems (LISs) are not equipped to track, collect, and display the different results in an effective and informative manner to support this workflow.
The aim of this project was to design and implement a web-based software application that centralizes test results for pathologist review, performs the quality control (QC) calculations for the complex LC-MS/MS analysis of opioids and their metabolites, functions as a user input form for pathologist interpretation entry, and auto-files results into the LIS.
Methods
The initial step before designing the application was to understand the entire workflow, then gather requirements and specifications from stakeholders.
Pre-application testing and workflows
At UW Medicine, for each patient specimen undergoing the opiate confirmation assay, two extractions are prepared, an undiluted and a diluted 10-fold urine sample. To help improve the complex LC-MS/MS data analysis of opioids and opioid metabolites, a command-line software application was developed. [10] The software application known as “SMACK” was shown to improve the data analysis process by automating a QC algorithm and calculations to improve consistency of analysis as well as reduce the amount of time medical laboratory scientists (MLS) spent reviewing the data.
The algorithm was part of an intricate workflow that involved several steps, spreadsheets, and individuals. The goal of the spreadsheets was to collect and consolidate the relevant patient information so that the reviewing pathologist had the necessary information to generate an interpretation for the overall case. The information with the relevant patient data was spread across several spreadsheets, the creation of which relied on laboratory staff and pathologists copying and pasting various fields from one spreadsheet to another. Pathologists and laboratory staff communicated the completion of steps via email while using a naming convention for the relevant files.
Requirements
We set to gather information on how clinicians ordered the different drug monitoring tests and how the information arrived at the laboratory. We investigated the LIS data workflow that was in place and how this information could feed the application, including assay results and patient demographics. Additionally, we worked with the laboratory to understand how test results were produced and how this information should be included in the application. Lastly, we investigated what the requirements were for progressing from one step of the workflow to the next along with the data associated with the progression. In addition to the workflow in the laboratory, we needed to understand what data elements are needed for each step of the workflow. With this information, we would create a map of the workflow to visualize and better understand the workflow. We gathered necessary components from stakeholders, pathologists, directors, pathologists in training, and laboratory staff by interviewing stakeholders and observing workflows. During this process, we gathered user interface requirements from stakeholders to shape functionality of the optimized web application.
Technical requirements
The previously described QC software SMACK had been implemented as a command-line application in Python, with no dependencies outside of the standard library. [10] Input data is provided as an XML format file exported from Waters' TargetLynx software. The outputs of the software are two CSV-formatted reports, one containing assay results and the other a detailed report of the QC calculations. Appendix A, Supplemental Fig. 1 displays the flow chart defining the QC algorithm, which has been simplified since its initial implementation. For this project, we needed to incorporate the software into the application to perform the same QC calculations on the opiate assay raw data and then display the output to the user in a meaningful manner. Moreover, our group has a defined technical software stack for developing and deploying applications in Python. For these reasons, writing the application in Python was the most efficient choice. An additional requirement was to automatically file results into the LIS (from CliniSys, Inc.). To achieve this requirement, we had to work with Data Innovations' Instrument Manager software.
Performance measurement and user feedback
Finally, to assess the performance, we surveyed stakeholders and documented the rough amount of time that is required in each step of the workflow. Input was solicited from several laboratory staff members rotating through the area regarding how much time was spent on each step before and after the implementation of the application. We also surveyed the pathologists in training (chemistry fellows and rotating residents), known as trainees. Finally, we extracted data from the LIS to capture the number of occurrences in which a testing order had a correction or modification after the initial data entry so we could compare correction rates prior to and after implementation.
Results
Laboratory workflows prior to implementation
Providers order UDTs for the patient based on an assessment of the patient's risk for compliance. Table 1 displays the patient risk levels, the intended population, and the tests associated with each risk panel.
| Table 1. Urine drug testing ordering strategy based on risk. Providers assess risk, review clinical patient care data, and order tests as described. | ||
| Risk | Intended population | Tests |
|---|---|---|
| Low | Morphine equivalent dose <90 mg/day; Stable on chronic opioid therapy or buprenorphine therapy | Drug screen immunoassay |
| Medium to high | Morphine equivalent dose >=90 mg/day; Aberrant behavior (including early prescriptions, lost prescriptions, outbursts in office) | Drug screen immunoassay; Enzymatic assay for alcohol; Opiate confirmation LCMS |
| High | Same as medium to high; added concern for clonazepam or lorazepam | Drug screen immunoassay; Enzymatic assay for alcohol; Opiate confirmation LCMS; Benzodiazepine confirmation LCMS |
For each panel, providers have the option to enter any drug the patient is expected to be taking which may influence the test results. Each test in the panel is conducted separately and the results are entered into the LIS. Once all the tests have been completed and results entered, the pathologist may begin generating an interpretation.
When pathologists are reviewing the results, they consider the patient's history in the electronic health records (EHRs) for information that may influence the test results such as medications not specified by the ordering provider. These notes would be entered in dedicated fields in the spreadsheet. For training purposes, a pathologist trainee will often review the results first, then review their work with a director who may discuss the case, edit the preliminary interpretation, then make the final approval. Upon review of cases that initially only received the immunoassay drug screen, a probable outcome is for the reviewing pathologist to deem additional confirmation testing necessary. Therefore, the workflow is the summation of two separate parts, cases which receive opiate confirmation testing versus those that do not. The consideration to separate the workflow into two parts was partly because many orders require additional reflexive orders which influences how quickly the final interpretations will be released. For this reason, it was best to group cases by risk level to provide an interpretation as quickly as possible. Laboratory staff worked with departmental software engineers to create and deliver spreadsheets for each workflow which contain the patient's demographics (medical record number, age, sex, name, and order number) along with the results of the tests.
The opiate assay is performed on a Waters Xevo TQ-MS tandem mass spectrometer with an Aquity UPLC system. Data acquisition is controlled by Waters MassLynx and chromatography review is completed using Waters TargetLynx software. When the data acquisition is complete, the technologist completes a cursory review of the auto-integration using TargetLynx. Once the preliminary review is complete, an XML file with the raw data is created. The XML file is the input of the application that performs QC calculations from the LC-MS/MS raw data. The output was two detailed tabular reports; the first was the calculated results after the algorithm was applied and another with a detailed report of the QC calculations. These reports would be reviewed in Microsoft Excel by two laboratory staff members to address any values that did not meet QC standards before entering the results into the LIS. Once the results have been reviewed, staff would send an email to the reviewing pathologists that reports were ready for their review. The pathologist would then combine the calculated results with the spreadsheet containing other test results and patient demographics via a copy-paste method. During the pathologist review, they may identify aberrant results stemming from interfering compounds or possible adulterations based on the data. They would then create an email thread with the laboratory to communicate and resolve the issue.
One possible outcome for those cases where patient risk levels are associated with low to medium risk, a reviewing pathologist orders a confirmatory test. Pathologists would type the test codes for the orders they wished to place in the spreadsheet and the reviewing MLS would place the orders in the LIS. Therefore, a third spreadsheet was created to capture the results for the cases that received additional tests. To transfer the notes captured during the initial review, a Microsoft Excel macro was created which would parse the spreadsheets and bring the pathologist's saved work from the initial spreadsheet into the final spreadsheet.
These spreadsheets were created and delivered via a dedicated website where staff and faculty were able to download the spreadsheets from any computer in the laboratory. After the spreadsheet was initially downloaded and edited, the file was stored on a shared drive accessible through a protected network that was accessible to the laboratory and pathologists.
A naming convention for these files was created to help indicate the stage in the workflow.
Once the reviewing pathologist had completed their review, they would send an email to staff so that they could enter the interpretation result into the LIS manually. If the technologist identified any issues with the interpretation, they would email the pathologist to edit/review the interpretation.
Technical requirements
The application was implemented in Python using the Flask web framework[1] and other open-source libraries. Appendix A, Supplemental Table 1 lists the required libraries used for the application.
The software was developed on an Apple Macintosh running Mac OS 10.15 and is used in production on Amazon Web Services (AWS) server running Ubuntu (18.04 LTS). The data from the LIS is gathered using Cache Object Script that collects information every 30 minutes from 6 a.m. to 6 p.m. and is delivered to a secure cloud storage resource (S3 bucket) on AWS. Once the results are finalized, the application sends a tab-delimited file to a transfer Linux (4.15.0) server which is picked up by Data Innovations instrument manager that sends the data to the LIS.
An important component of the software development and deployment process was the use of the Git version control software. Git is a free, open-source, distributed version control software that can be used to store “snapshots” of source code files in which modifications are captured to the code repository. Each change is attributed to a specific author and can be associated with a comment describing the intent of the change. A unique tag identifies each change, and the application captures the tag in the software version number and records this identifier to every data output. In this manner, results of each analysis can be traced to the exact version of the software used to collect and generate those results. Additionally, GitLab is a Git-based fully integrated platform which allows users to create “issues” where enhancements, development, and other topics may be discussed collectively. This feature of GitLab can also attribute labels to each issue so that larger topics or concepts can be collectively viewed and labeled.
The application is a web-based application that can be launched from any web browser. Access control is managed using our institutional identity provider with single sign-on (SSO) with two-factor authentication (2FA). For auto-resulting, once the results reach their terminal status (pathologists and laboratory staff have completed their collective review), a tab-separated-values (TSV) is sent to Data Innovations' Instrument Manager where it files the results to the LIS.
Requirements
The data collected and workflows described serviced as the foundation of the application's design. The application needed to gather the necessary data from the LIS and display the information in a manner that is useful to the user. A system needed to be designed to progress a case through the necessary steps for the appropriate reviewer. Entry fields needed to be available for all members to be able to communicate problems, capture notes by the pathologists during their review, and for the final interpretation result. The application needed to ingest the XML file from TargetLynx of the opiate test, apply the SMACK algorithm, display both the QC calculations and results, and save all edits. Lastly, the application needed a system to communicate any additional confirmation tests the pathologist wanted to order.
A new component within the laboratory workflow was the introduction of an automated liquid handler to prepare the urine samples for the opiate assay. The laboratory had been manually preparing the samples and entering the patient identifiers into the acquisition system, MassLynx. When creating the extraction method of the liquid handler, the decision was made to use the barcode on the specimen label as the sample identifier for the assay. To associate the result back to the patient, all specimen IDs also needed to be gathered.
The decision to create a web-based application was to provide an easy-access entry point for the laboratory and pathologists. Also, a web-based application provided the flexibility needed to create a custom site that fit the needs of the workflow. Fig. 1 shows the home page of the application.
|
References
- ↑ "Flask". Pallets. 2010. https://flask.palletsprojects.com/en/2.3.x/.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation, spelling, and grammar. In some cases important information was missing from the references, and that information was added.