Journal:Data management and modeling in plant biology
| Full article title | Data management and modeling in plant biology |
|---|---|
| Journal | Frontiers in Plant Science |
| Author(s) | Krantz, Maria; Zimmer, David; Adler, Stephan O.; Kitashova, Anastasia; Klipp, Edda; Mühlhaus, Timo; Nägele, Thomas |
| Author affiliation(s) | Humboldt-Universität zu Berlin, Technische Universität Kaiserslautern, Ludwig-Maximilians-Universität München |
| Primary contact | Email: thomas dot naegele at lmu dot de |
| Editors | Fukushima, Atsushi |
| Year published | 2021 |
| Volume and issue | 12 |
| Article # | 717958 |
| DOI | 10.3389/fpls.2021.717958 |
| ISSN | 1664-462X |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.frontiersin.org/articles/10.3389/fpls.2021.717958/full |
| Download | https://www.frontiersin.org/articles/10.3389/fpls.2021.717958/pdf (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
The study of plant-environment interactions is a multidisciplinary research field. With the emergence of quantitative large-scale and high-throughput techniques, the amount and dimensionality of experimental data have strongly increased. Appropriate strategies for data storage, management, and evaluation are needed to make efficient use of experimental findings. Computational approaches to data mining are essential for deriving statistical trends and signatures contained in data matrices. Although, current biology is challenged by high data dimensionality in general, this is particularly true for plant biology. As sessile organisms, plants have to cope with environmental fluctuations. This typically results in strong dynamics of metabolite and protein concentrations, which are often challenging to quantify. Summarizing experimental output results in complex data arrays, which need computational statistics and numerical methods for building quantitative models. Experimental findings need to be combined with computational models to gain a mechanistic understanding of plant metabolism. For this, bioinformatics and mathematics need to be combined with experimental setups in physiology, biochemistry, and molecular biology. This review presents and discusses concepts at the interface of experiment and computation, which are likely to shape current and future plant biology. Finally, this interface is discussed with regard to its capabilities and limitations to develop a quantitative model of plant-environment interactions.
Keywords: genome-scale networks, omics analysis, metabolic regulation, plant-environment interactions, machine learning, mathematical modeling, differential equations
Introduction
Experimental high-throughput analysis of genomes, transcriptomes, proteomes, and metabolomes results in a vast number of simultaneously quantified molecular entities. Current biological research frequently applies a combination of experimental high-throughput techniques to address a wide spectrum of complex research questions. On the genome level, high-throughput sequencing (HTS) technologies have revolutionized genetics and genomics, and sequencing projects have provided comprehensive information about many species’ genomes.[1][2][3][4][5] To date, thousands of genomes have been sequenced and pan-genomics approaches have been initiated, which assemble diverse sets of individual genomes to a collection of all DNA sequences occurring in a species.[6] In plant sciences, the concept of pan-genomics is already discussed to support breeding strategies or evolutionary studies and may significantly contribute to the explanation of gene presence and absence variation.[7]
Based on such comprehensive genome information, genome-scale models of plant metabolism have been developed and applied to predict plant metabolism in a diverse context. Validation and biotechnological application of such large-scale models need appropriate experimental techniques and platforms, unifying sample analysis in multi-omics approaches.[8] Although, omics techniques have become a generic element of numerous research projects to quantify transcripts, proteins, and metabolites, the actual handling, normalization, and integration of multidimensional experimental data output is still a central challenge in biology.[9] The need for integrative analysis of experimental high-throughput data has already been suggested and discussed earlier. For example, almost a decade ago, integrative approaches were suggested for transcriptomics, proteomics, and metabolomics data to promote a systems-level understanding of the genus Arabidopsis.[10] Since then, machine learning, computational statistics, and mathematical modeling have significantly advanced data integration strategies. Due to their capability to improve the understanding of the genotype-phenotype relation on a molecular level, systems biology, and multi-omics integration have become central topics in the discussion about future perspectives of biology and medicine. Yet, in order to make experiments comparable and to increase consistency and reproducibility across different experimental platforms, laboratories, or research communities, quantitative omics data are needed.[11] Furthermore, quantitative experimental data necessitates appropriate processing strategies to make it comparable to other independent studies and statistics. Making data and data processing publicly available via databases and repositories may represent one of the most important steps to establish and expand a cross-disciplinary scientific platform for omics data integration. Together with the need for traceable long-term data storage and versioning, these topics are becoming increasingly important in quantitative biology.
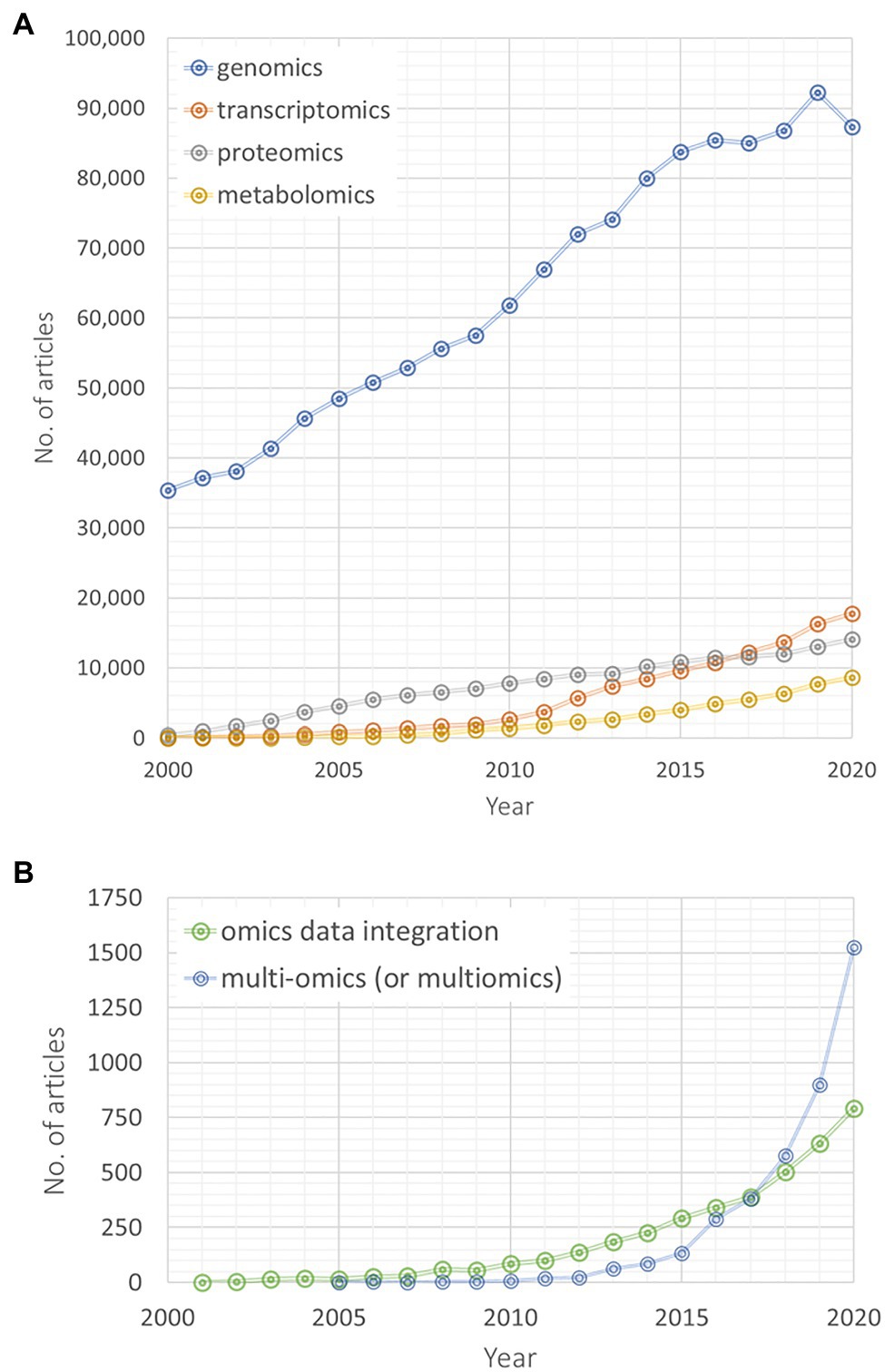
Searching for database entries from the last two decades on omics and integrative omics approaches reveals a rapidly increasing research and publication activity in the integrative multi-omics research field (Figure 1). Genomics-related yearly published articles linearly increased to a very high level during the last 20 years, while particularly transcriptomics and metabolomics articles have been published with an increasing rate during the last decade (Figure 1A). Between 2000 and 2015, more proteomics-related articles have been published than transcriptomics and metabolomics articles, but since 2017 their number lies between both omics disciplines. Interestingly, since 2017, articles searchable by the queries “multi-omics” or “multiomics” are exponentially increasing in their number (Figure 1B). A similar, yet weaker trend is also observable for “omics data integration” articles (Figure 1B). Of course, these numbers are only crude estimates based on our chosen specific vocabulary and searched within one specific database (for example, we have not checked the combination of different omics disciplines, i.e., “genomics” and “transcriptomics” instead of “multi-omics”). Yet, these results still indicate that an increasing number of studies focuses on a multi-omics design and that omics data integration gains more and more attention.
|
This article aims to summarize and discuss current advances and limitations of integrative molecular analysis, computational modeling, and data science. It focuses on both experimental and theoretical methodology to support design and analysis of interdisciplinary research in plant biology. A particular focus is laid on methodologies for capturing system dynamics of plant metabolism induced by a changing environment.
On a large scale: How does genome-scale metabolic network reconstruction support data integration in plant biology?
The availability of comprehensive genome information has enabled the reconstruction of genome-scale metabolic networks, which predict, based on gene annotation, a functional cellular network structure. This crucially supports the interpretation of gene functions and makes pathways accessible to computational biology and mathematics.[12] Further, reconstructed networks significantly facilitate a mechanistic description of genotype-phenotype relationships and enable the application of constraint-based analysis methods.[13][14] Major constraints are thermodynamics, mass and charge conservation, and the substrate/enzyme availability. Constraints dramatically reduce the parameter space, which explains a genotype-phenotype relationship, and, hence, strongly increases the probability to find physiologically relevant solutions for underlying equation systems. Thus, it is not surprising that, in current plant biology, genome-scale reconstruction has become an integral part from single-cell to multi-tissue modeling.[15] For example, model reconstructions have been applied to analyze metabolic regulation in autotrophic and heterotrophic tissues, to study C4 plant metabolism, to evaluate diurnal metabolic interactions in plant leaf tissue and to analyze photorespiration.[16][17][18][19]
The experimental basis for constraining, validating, and optimizing large-scale models are high-throughput experiments, i.e., omics analyses. For example, to investigate effects of nitrogen assimilation on metabolism in maize (Zea mays), a genome-scale metabolic model for maize leaf was created comprising more than 5,800 genes, 8,500 reactions, and 9,000 metabolites.[20] Using a combination of transcriptomic and proteomic data to constrain metabolic flux predictions, the authors were able to reproduce experimentally determined metabolomic data to significantly higher accuracy than without these constraints. Applying a combination of publicly available data on maize metabolism, reaction networks, and results from omics experiments, information about reaction stoichiometry, directionality, and compartmentalization was derived. Algorithmic model curation was combined with manual modification to, for example, resolve gaps in the network model with reactions from similar organisms. Information about transcripts and proteins, which were experimentally observed to significantly differ in mutants and under variable nitrogen supply, were then incorporated into the model by switching on/off corresponding reactions. Flux predictions through the metabolic network were compared to metabolomics measurements. With this integrated setup, model application unraveled genes coding for enzymes, which are involved in regulation of biomass formation under variable nitrogen supply.[20] In another study, publicly available transcriptomics and metabolomics data were used within a constraint-based modeling approach to investigate network structure and flux distribution in root cell types and tissue layers of Arabidopsis thaliana. Based on transcriptomics and metabolomics data, it was possible to extract tissue and cell type specific models from a general genome-scale model of root metabolism. By this, the authors were able to simulate and analyze cell types as autonomous subsystems, which communicate with each other via metabolites or proteins. But it was also shown and discussed that further experimental evidence and constraints are essential to support hypotheses derived from their simulations.[21] This example nicely illustrates how large-scale data integration can (i) unravel novel and detailed mechanistic insights into plant metabolism, and also (ii) indicate design and research focus of follow-up studies to prove model predictions. By placing metabolites, proteins, or transcripts into a pathway and network context, genome-scale models significantly support the biochemical and physiological interpretation of molecular data.
Also, in a biotechnological context, such data integration strategies have become an important and promising tool to advance and improve bioengineering strategies. As an example, a genome-scale metabolic network reconstruction for green microalgal model species Chlamydomonas reinhardtii has been developed which reliably and quantitatively predicts growth depending on the light source.[22] This metabolic network comprises 10 compartments, accounting for more than 1,000 genes associated with more than 2,000 reactions and over 1,000 metabolites. Regulatory effects arising from different light conditions are covered by the model, which enables estimation of growth under different laboratory conditions. The model has been refined using metabolite profiling to include further branches of metabolism, e.g., amino acids and peptides as nitrogen sources.[23] Although, it developed a decade ago, the original model (named iRC1080) still represents a valid and supportive platform for data interpretation, and it still fruitfully initiates further model development and validation.[24] These examples, together with many other studies that have been summarized recently[25], provide strong evidence for the capability of genome-scale metabolic models to couple statistics with metabolic models.
References
- ↑ International Human Genome Sequencing Consortium; Whitehead Institute for Biomedical Research, Center for Genome Research:; Lander, Eric S.; Linton, Lauren M.; Birren, Bruce; Nusbaum, Chad; Zody, Michael C.; Baldwin, Jennifer et al. (15 February 2001). "Initial sequencing and analysis of the human genome" (in en). Nature 409 (6822): 860–921. doi:10.1038/35057062. ISSN 0028-0836. http://www.nature.com/articles/35057062.
- ↑ The 1000 Genomes Project Consortium (1 November 2012). "An integrated map of genetic variation from 1,092 human genomes" (in en). Nature 491 (7422): 56–65. doi:10.1038/nature11632. ISSN 0028-0836. PMC PMC3498066. PMID 23128226. http://www.nature.com/articles/nature11632.
- ↑ Alonso-Blanco, Carlos; Andrade, Jorge; Becker, Claude; Bemm, Felix; Bergelson, Joy; Borgwardt, Karsten M.; Cao, Jun; Chae, Eunyoung et al. (1 July 2016). "1,135 Genomes Reveal the Global Pattern of Polymorphism in Arabidopsis thaliana" (in en). Cell 166 (2): 481–491. doi:10.1016/j.cell.2016.05.063. PMC PMC4949382. PMID 27293186. https://linkinghub.elsevier.com/retrieve/pii/S0092867416306675.
- ↑ Stein, Joshua C.; Yu, Yeisoo; Copetti, Dario; Zwickl, Derrick J.; Zhang, Li; Zhang, Chengjun; Chougule, Kapeel; Gao, Dongying et al. (1 February 2018). "Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza" (in en). Nature Genetics 50 (2): 285–296. doi:10.1038/s41588-018-0040-0. ISSN 1061-4036. http://www.nature.com/articles/s41588-018-0040-0.
- ↑ Sun, Hequan; Rowan, Beth A.; Flood, Pádraic J.; Brandt, Ronny; Fuss, Janina; Hancock, Angela M.; Michelmore, Richard W.; Huettel, Bruno et al. (1 December 2019). "Linked-read sequencing of gametes allows efficient genome-wide analysis of meiotic recombination" (in en). Nature Communications 10 (1): 4310. doi:10.1038/s41467-019-12209-2. ISSN 2041-1723. PMC PMC6754367. PMID 31541084. http://www.nature.com/articles/s41467-019-12209-2.
- ↑ Sherman, Rachel M.; Salzberg, Steven L. (1 April 2020). "Pan-genomics in the human genome era" (in en). Nature Reviews Genetics 21 (4): 243–254. doi:10.1038/s41576-020-0210-7. ISSN 1471-0056. PMC PMC7752153. PMID 32034321. http://www.nature.com/articles/s41576-020-0210-7.
- ↑ Bayer, Philipp E.; Golicz, Agnieszka A.; Scheben, Armin; Batley, Jacqueline; Edwards, David (1 August 2020). "Plant pan-genomes are the new reference" (in en). Nature Plants 6 (8): 914–920. doi:10.1038/s41477-020-0733-0. ISSN 2055-0278. http://www.nature.com/articles/s41477-020-0733-0.
- ↑ Weckwerth, Wolfram; Ghatak, Arindam; Bellaire, Anke; Chaturvedi, Palak; Varshney, Rajeev K. (1 July 2020). "PANOMICS meets germplasm" (in en). Plant Biotechnology Journal 18 (7): 1507–1525. doi:10.1111/pbi.13372. ISSN 1467-7644. PMC PMC7292548. PMID 32163658. https://onlinelibrary.wiley.com/doi/10.1111/pbi.13372.
- ↑ Scossa, Federico; Alseekh, Saleh; Fernie, Alisdair R. (1 February 2021). "Integrating multi-omics data for crop improvement" (in en). Journal of Plant Physiology 257: 153352. doi:10.1016/j.jplph.2020.153352. https://linkinghub.elsevier.com/retrieve/pii/S017616172030242X.
- ↑ Liberman, Louisa M; Sozzani, Rosangela; Benfey, Philip N (1 April 2012). "Integrative systems biology: an attempt to describe a simple weed" (in en). Current Opinion in Plant Biology 15 (2): 162–167. doi:10.1016/j.pbi.2012.01.004. PMC PMC3435099. PMID 22277598. https://linkinghub.elsevier.com/retrieve/pii/S1369526612000052.
- ↑ Pinu, Farhana R.; Beale, David J.; Paten, Amy M.; Kouremenos, Konstantinos; Swarup, Sanjay; Schirra, Horst J.; Wishart, David (18 April 2019). "Systems Biology and Multi-Omics Integration: Viewpoints from the Metabolomics Research Community" (in en). Metabolites 9 (4): 76. doi:10.3390/metabo9040076. ISSN 2218-1989. PMC PMC6523452. PMID 31003499. https://www.mdpi.com/2218-1989/9/4/76.
- ↑ Oberhardt, Matthew A; Palsson, Bernhard Ø; Papin, Jason A (1 January 2009). "Applications of genome‐scale metabolic reconstructions" (in en). Molecular Systems Biology 5 (1): 320. doi:10.1038/msb.2009.77. ISSN 1744-4292. PMC PMC2795471. PMID 19888215. https://onlinelibrary.wiley.com/doi/10.1038/msb.2009.77.
- ↑ Lewis, Nathan E.; Nagarajan, Harish; Palsson, Bernhard O. (1 April 2012). "Constraining the metabolic genotype–phenotype relationship using a phylogeny of in silico methods" (in en). Nature Reviews Microbiology 10 (4): 291–305. doi:10.1038/nrmicro2737. ISSN 1740-1526. PMC PMC3536058. PMID 22367118. http://www.nature.com/articles/nrmicro2737.
- ↑ Ramon, Charlotte; Gollub, Mattia G.; Stelling, Jörg (26 October 2018). "Integrating –omics data into genome-scale metabolic network models: principles and challenges" (in en). Essays in Biochemistry 62 (4): 563–574. doi:10.1042/EBC20180011. ISSN 0071-1365. https://portlandpress.com/essaysbiochem/article/62/4/563/78519/Integrating-omics-data-into-genome-scale-metabolic.
- ↑ Gomes de Oliveira Dal’Molin, Cristiana; Nielsen, Lars Keld (1 February 2018). "Plant genome-scale reconstruction: from single cell to multi-tissue modelling and omics analyses" (in en). Current Opinion in Biotechnology 49: 42–48. doi:10.1016/j.copbio.2017.07.009. https://linkinghub.elsevier.com/retrieve/pii/S0958166917301052.
- ↑ de Oliveira Dal’Molin, Cristiana Gomes; Quek, Lake-Ee; Palfreyman, Robin William; Brumbley, Stevens Michael; Nielsen, Lars Keld (1 December 2010). "C4GEM, a Genome-Scale Metabolic Model to Study C4 Plant Metabolism" (in en). Plant Physiology 154 (4): 1871–1885. doi:10.1104/pp.110.166488. ISSN 1532-2548. PMC PMC2996019. PMID 20974891. https://academic.oup.com/plphys/article/154/4/1871/6108787.
- ↑ de Oliveira Dal'Molin, Cristiana Gomes; Quek, Lake-Ee; Palfreyman, Robin William; Brumbley, Stevens Michael; Nielsen, Lars Keld (3 February 2010). "AraGEM, a Genome-Scale Reconstruction of the Primary Metabolic Network in Arabidopsis" (in en). Plant Physiology 152 (2): 579–589. doi:10.1104/pp.109.148817. ISSN 1532-2548. PMC PMC2815881. PMID 20044452. https://academic.oup.com/plphys/article/152/2/579/6108441.
- ↑ Cheung, C.Y. Maurice; Poolman, Mark G.; Fell, David. A.; Ratcliffe, R. George; Sweetlove, Lee J. (2 June 2014). "A Diel Flux Balance Model Captures Interactions between Light and Dark Metabolism during Day-Night Cycles in C3 and Crassulacean Acid Metabolism Leaves" (in en). Plant Physiology 165 (2): 917–929. doi:10.1104/pp.113.234468. ISSN 1532-2548. PMC PMC4044858. PMID 24596328. https://academic.oup.com/plphys/article/165/2/917/6113238.
- ↑ Yuan, Huili; Cheung, C.Y. Maurice; Poolman, Mark G.; Hilbers, Peter A. J.; Riel, Natal A. W. (1 January 2016). "A genome‐scale metabolic network reconstruction of tomato ( Solanum lycopersicum L.) and its application to photorespiratory metabolism" (in en). The Plant Journal 85 (2): 289–304. doi:10.1111/tpj.13075. ISSN 0960-7412. https://onlinelibrary.wiley.com/doi/10.1111/tpj.13075.
- ↑ 20.0 20.1 Simons, Margaret; Saha, Rajib; Amiour, Nardjis; Kumar, Akhil; Guillard, Lenaïg; Clément, Gilles; Miquel, Martine; Li, Zhenni et al. (5 November 2014). "Assessing the Metabolic Impact of Nitrogen Availability Using a Compartmentalized Maize Leaf Genome-Scale Model" (in en). Plant Physiology 166 (3): 1659–1674. doi:10.1104/pp.114.245787. ISSN 1532-2548. PMC PMC4226342. PMID 25248718. https://academic.oup.com/plphys/article/166/3/1659/6111218.
- ↑ Scheunemann, Michael; Brady, Siobhan M.; Nikoloski, Zoran (1 December 2018). "Integration of large-scale data for extraction of integrated Arabidopsis root cell-type specific models" (in en). Scientific Reports 8 (1): 7919. doi:10.1038/s41598-018-26232-8. ISSN 2045-2322. PMC PMC5962614. PMID 29784955. http://www.nature.com/articles/s41598-018-26232-8.
- ↑ Chang, Roger L; Ghamsari, Lila; Manichaikul, Ani; Hom, Erik F Y; Balaji, Santhanam; Fu, Weiqi; Shen, Yun; Hao, Tong et al. (1 January 2011). "Metabolic network reconstruction of Chlamydomonas offers insight into light‐driven algal metabolism" (in en). Molecular Systems Biology 7 (1): 518. doi:10.1038/msb.2011.52. ISSN 1744-4292. PMC PMC3202792. PMID 21811229. https://onlinelibrary.wiley.com/doi/10.1038/msb.2011.52.
- ↑ Chaiboonchoe, Amphun; Dohai, Bushra Saeed; Cai, Hong; Nelson, David R.; Jijakli, Kenan; Salehi-Ashtiani, Kourosh (10 December 2014). "Microalgal Metabolic Network Model Refinement through High-Throughput Functional Metabolic Profiling". Frontiers in Bioengineering and Biotechnology 2. doi:10.3389/fbioe.2014.00068. ISSN 2296-4185. PMC PMC4261833. PMID 25540776. http://journal.frontiersin.org/article/10.3389/fbioe.2014.00068/abstract.
- ↑ Shene, Carolina; Asenjo, Juan A.; Chisti, Yusuf (1 December 2018). "Metabolic modelling and simulation of the light and dark metabolism of Chlamydomonas reinhardtii" (in en). The Plant Journal 96 (5): 1076–1088. doi:10.1111/tpj.14078. https://onlinelibrary.wiley.com/doi/10.1111/tpj.14078.
- ↑ Tong, Hao; Nikoloski, Zoran (1 February 2021). "Machine learning approaches for crop improvement: Leveraging phenotypic and genotypic big data" (in en). Journal of Plant Physiology 257: 153354. doi:10.1016/j.jplph.2020.153354. https://linkinghub.elsevier.com/retrieve/pii/S0176161720302443.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation, spelling, and grammar. In some cases important information was missing from the references, and that information was added. The original article lists references in alphabetical order; however, this version lists them in order of appearance, by design.