Journal:FAIR Health Informatics: A health informatics framework for verifiable and explainable data analysis
| Full article title | FAIR Health Informatics: A health informatics framework for verifiable and explainable data analysis |
|---|---|
| Journal | Healthcare |
| Author(s) | Siddiqi, Muhammad H.; Idris, Muhammad; Alruwaili, Madallah |
| Author affiliation(s) | Jouf University, Universite Libre de Bruxelles |
| Primary contact | Email: mhsiddiqi at ju dot edu dot sa |
| Year published | 2023 |
| Volume and issue | 11(12) |
| Article # | 1713 |
| DOI | 10.3390/healthcare11121713 |
| ISSN | 2227-9032 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.mdpi.com/2227-9032/11/12/1713 |
| Download | https://www.mdpi.com/2227-9032/11/12/1713/pdf?version=1686475395 (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
The recent COVID-19 pandemic has hit humanity very hard in ways rarely observed before. In this digitally connected world, the health informatics and clinical research domains (both public and private) lack a robust framework to enable rapid investigation and cures. Since data in the healthcare domain are highly confidential, any framework in the healthcare domain must work on real data, be verifiable, and support reproducibility for evidence purposes. In this paper, we propose a health informatics framework that supports data acquisition from various sources in real-time, correlates these data from various sources among each other and to the domain-specific terminologies, and supports querying and analyses. Various sources include sensory data from wearable sensors, clinical investigation (for trials and devices) data from private/public agencies, personal health records, academic publications in the healthcare domain, and semantic information such as clinical ontologies and the Medical Subject Headings (MeSH) ontology. The linking and correlation of various sources include mapping personal wearable data to health records, clinical oncology terms to clinical trials, and so on. The framework is designed such that the data are findable, accessible, interoperable, and reusable (FAIR) with proper identity and access management mechanisms. This practically means tracing and linking each step in the data management lifecycle through discovery, ease of access and exchange, and data reuse. We present a practical use case to correlate a variety of aspects of data relating to a certain medical subject heading from the MeSH ontology and academic publications with clinical investigation data. The proposed architecture supports streaming data acquisition, and servicing and processing changes throughout the lifecycle of the data management process. This is necessary in certain events, such as when the status of a certain clinical or other health-related investigation needs to be updated. In such cases, it is required to track and view the outline of those events for the analysis and traceability of the clinical investigation and to define interventions if necessary.
Keywords: data correlation, data linking, verifiable data, data analysis, explainable decisions, clinical trials, COVID, clinical investigation, semantic mapping, smart health
Introduction
Pandemics are not new to this world or humanity. There have been pandemics in the past, and they may happen again in the future. The recent COVID-19 pandemic is different from the previous ones in that the virus is more infectious without being known, symptoms are ambiguous, and the detection methods require a lot of time and resources. It has caused more deaths than ever in the history of humankind, and the impact it has had on the world economy and human lives (whether affected or not) is grave and is posing questions about the future of diseases and pandemics. At the same time, while humans advance knowledge and technology, there is a need to investigate and put effort into overcoming the challenges posed by these kinds of serious threats. This not only requires us to deal with the current pandemic but also to look into the future, predict and presume the possibilities, investigate, and develop solutions at a large scale so that there is a reduction in the risk of losing lives and danger on a large scale.
The majority of the existing software systems and solutions in the healthcare domain are proprietary and limited in their capacity to a specific domain, such as only processing clinical trial data without integrating state-of-the-art investigation and wearable sensor data. Hence, they lack the ability to present a scalable analytical and technical solution, while having limitations and lacking the ability to trace back the analysis and results to the origin of the data. Because of these limitations, any robust and practical solution does not only have to account for clinical data but also present a practical and broader overview of these catastrophic events from clinical investigations, their results and treatments that are up-to-date, and combine them with medical history records as well as academic and other related datasets available. In other words, the recent advancements and investigations in the clinical domain specific to a particular disease are published in research articles and journals, and they also need to be correlated to real human subjects that undergo clinical trials so that up-to-date analysis can be carried out and proper guidelines and interventions can be suggested.
Moreover, the majority of the record-keeping bodies maintain electronic health records (EHRs) for patients and, recently, records of COVID vaccinations. However, they lack the ability to link the data to individuals’ activities and clinical outcomes. There is also a lack of fusing data related to the investigation of a particular disease from various data providers, such as private clinical trials, public clinical trials [1,2,3], and public-private clinical trials. The lack of these services is not only because there is less literature on fusing these multiple forms of data but also because of security and privacy concerns related to the confidentiality of healthcare data. The current advanced and robust privacy and security infrastructure available is more than enough to ensure personal and organizational interests. On the one hand, there are governments and other organizations that publish their clinical research data to public repositories to be available for clinical research using defined standards. On the other hand, there are pharma companies, which mostly hold the analytics driven by these and the respective algorithms and methods private. Furthermore, clinical trial data are not enough since they only provide measurements for different subjects who underwent a trial, while other data, from sources such as EHRs, contain real investigative cases and the histories of patients. Therefore, fusing these data from a variety of sources is helpful to determine the effects of a particular drug or treatment plan in combination with other vaccines, treatments, etc.
Given the above brief overview of the capabilities of the state-of-the-art, most of these systems either tackle static or dynamic, relational or non-relational, noisy or cleaned data without fusing, integrating, or semantically linking it. This paper proposes a solution to the above problems by presenting a healthcare framework that supports the ability to acquire, manage, and process static and dynamic (real-time) data. Our proposed framework is a data format that focuses on fusing information from various sources. In a nutshell, the proposed framework ideally targets a strategy that makes data findable, accessible, interoperable, and reusable (FAIR).
Objectives and contributions
In particular, we define the objectives and contributions of this research work as proposing a framework that is designed to be able to provide the following basic and essential capabilities for healthcare data:
- A clinical "data lake" that stores data in a unified format where the raw data can be in any format loaded from a raw storage or streamed.
- Pipelines that support static and incremental data collection from raw storage to a clinical data lake and maintain a record of any change to the structure at the data level and at the schema level all the way to the raw data and raw data schema.
- A schema repository that versions the data when it changes its structure and enables forward and backward compatibility of the data throughout.
- Universal clinical schemas that can incorporate any clinically related concepts (such as clinical trials from any provider) and support flexibility.
- Processes to perform change data capture (CDC) as the data proceed down the pipeline towards the applications, i.e., incremental algorithms and methods to enable incremental processing of incoming data and keep a log of only the data of interest.
- Timelines for changing data for a specific clinical investigation from a clinical trial data element such as a trial, site, investigator, etc., and include the ability to stream the data to the application, while providing semantic linking and profiling of subjects in the data.
These capabilities are meant to provide evidence of data analysis (i.e., where does the analysis go back in terms of data), traces of changes, and a holistic view of various clinical investigations running simultaneously at different places related to a certain specific clinical investigation. An example of that is when the COVID-19 vaccine was being developed; it was necessary to be able to trace the investigation of various efforts by independent bodies in a single place where one could see the phases of clinical trials of vaccines, their outcomes, treatments, and even the investigation sites and investigators. This could not only result in better decision-making but also in putting resources in suitable places and better planning for future similar scenarios.
State of the art
Many works exist in the literature that deal with various aspects of clinical data, ranging from data management and analysis to interoperability, the meaning of big data in healthcare and its future course, the standardization of clinical data (especially clinical trials), and the correlation and analysis of data from various sources. The following sections present a brief overview of existing works in these various aspects.
Data management
Much work exists in data management in the healthcare domain. These include the design and analysis of clinical trials [5], big data in healthcare [6], and others. [7] A detailed survey of big data in healthcare by Bahri et al. [8] provides further insights. The crux of all the work in data management is to store data on a large scale and then be able to process it efficiently and quickly. However, this is not the only scope of this paper, since this paper does not only communicate and present information about data management (where we resemble the existing work) but also presents additional key features that distinguish our work from the existing work. Those distinguishing features are presented as contributions in the introduction section.
Data interoperability and standardization
Data interoperability and standardization are two other key aspects of healthcare data management and analysis. Unlike other traditional data management systems, healthcare data—in particular clinical data—require more robust, universally known, and recognized interoperable methods and standards because the data are critical to healthcare and the healthcare investigations and diagnoses that come with it. For example, Health Level 7 (HL7) standards focus on standardizing clinical trial terminologies across various stakeholders in the world. Multiple investigation research centers need to exchange results, outcomes, treatments, etc. to come to a common conclusion for certain diseases and treatments. Therefore, a high quality of standards, as explained by Schulz et al. [9] and Hussain et al. [10], exists, and ontological [11] representations have been defined to represent the data in an interoperable manner universally. There are a wide range of resources in this domain, and the readers can see further details in the survey by Brundaga et al. [12] Our research scope goes beyond this topic of standardization and interoperability and focuses on a more abstract level where all data from all types of providers in all standards can be brought together for analysis and be able to incorporate changes and evolve as the timelines of investigations evolve.
Data analysis
Like any other field of data analysis, clinical healthcare data has also been widely studied for analysis and preparation. This includes strategies for dealing with missing data [9], correlating data from various sources and generating recommendations for better healthcare [13], and the use of machine learning (ML) approaches for investigating the diagnosis of vaccines, e.g., COVID-19 vaccines. [14] Some examples of works related to this specific field are presented by Brundage et al. [12] and Majumder and Minko. [15] Moreover, we also find efforts that investigate supporting clinical decisions by clinicians in various fields, and these types of systems are generally referred to as clinical decision support systems (CDSSs). [16] These types of systems generally focus on generating recommendations using artificial intelligence (AI) and ML techniques. However, they lack the ability to discover, link, and provide an analytical view of the process of clinical trials [2] under investigation. These trials may still be under investigation and not yet complete. On the other hand, this research work leaves the part of analysis for a specific disease, diagnosis, treatment, etc. to the user of this solution and focuses on presenting a unified system where data from a variety of sources can be obtained at one place and be able to perform any kind of analysis such as ML, data preparation tasks, profiling, and recommendations [13] as listed in the contributions and uniqueness section of this paper. This research does provide a real-time and robust view of the processes to support fast and reliable clinical investigation while the data are not yet complete or an investigation has been completed.
Comparison of the proposed FAIR Health Informatics framework to the state of the art
The proposed FAIR Health Informatics framework differs from and supersedes the existing state of the art across several perspectives. Firstly, existing approaches either focus on data standardization for interoperability and exchange of clinical data specifically or act on the data in silos. Secondly, most of the analytical frameworks’ usage is only with specialized datasets, such as clinical trials only, EHRs only, or sensory data only. Thirdly, all the above-discussed systems do not capture the building timeline of events (changes) and mostly work with static data by loading periodic batches. The proposed framework in this paper addresses these problems by fusing data from various sources, building profiles for entities, maintaining changes in events over time for the entities of interest, and avoiding silos of analysis and computation. Moreover, the solution is designed to support streaming, batch, and static data.
Methodology
In this section, the terms and symbols used throughout this paper will be introduced first as preliminaries, and then the overall architecture of the framework will be presented.
Preliminaries
The terms used in this paper are of two types: those that describe entities or subjects for which a dataset is produced by a data provider, and those that describe or represent a process that relates to the steps a dataset undergoes. A process may involve subjects as its input or output, but a vice versa approach is not possible.
Terms
Entity: An entity, E, is a clinical concept, disease, treatment, or a human being related to data that can be collected, correlated to, and analyzed in combination with other entities. For example, a vaccine for COVID-19 under investigation is considered an entity. Humans under monitoring for a vaccine trial comprise an entity. Clinical concepts, diseases, treatments, devices, etc. are the types of entities that will be referred to as “entity being investigated,” whereas human beings on which the entities are observed are referred to as “entity being observed.”
Subject: A subject, S, is an entity (i.e., SE) for which a data item or a measurement is recorded. For example, a person is a human entity, and a vaccine is a clinical concept entity.
Subject types: A subject type is a subject for which data items can be recorded and can either be an entity being investigated or an entity being observed.
Domain: A domain, D, is a contextual entity and can be combined with a specific “entity being investigated” subject-type. For example, “breast cancer” is a domain, and "viral drug" is a domain. Moreover, in clinical terms, each high-level concept in Medical Subject Headings (MeSH) ontology [17] is a domain. Similarly, “electronic health record” (or "EHR") is a domain.
Sub-domain: Just like a subject with sub-types, a domain has sub-domains, e.g., “cancer” is a domain and “breast cancer” is a sub-domain. Furthermore, sub-domains can have further sub-domains.
Schema: A schema, Sch, is the template in which measurements or real values of a subject are recorded. A schema specifies the types, names, hierarchy, and arity of values in a measurement or a record. Schema is also referred to as type-level records.
Instance: An instance, I, is an actual record or a measurement that corresponds to a schema for the subjects of a particular domain. An instance of an EHR record belongs to a subject “person” of entity “human,” with entity type as an “entity being observed.” Similarly, a clinical trial record for “breast cancer” investigation with all its essential data (as described in coming sections) is an instance of entity “trial” in the domain “cancer” with sub-domain “breast cancer” and is an entity of type “entity being investigated.” Moreover, a set {I} of instances is referred to by a dataset, D, such that D has a schema, Sch.
Stakeholder: A stakeholder is a person, an organization, or any other such entity that needs to either onboard their data in the framework for analysis, use the framework with existing data for insights, or do both.
Scenario: A scenario, Sce, is a representation of a query that defines the parameters, domain, context, and scope of the intended use of the framework. For example, a possible scenario is when a stakeholder wants to visualize the timeline of investigations/trials for a certain “domain” (i.e., “breast cancer”) in the last two years by a particular investigation agency/organization. The scenario is then the encapsulation of all such parameters and context.
Use case: A use case, UC, is a practical scenario represented by steps and actions in a flow from the start of raw data until the point of analytical/processed data intended to show the result of the scenario, Sce.
Timeline: A timeline is normally the sequence of data-changing events in a particular entity being monitored. For example, in the case of a clinical trial, it is every change to any of its features, such as the number of registered patients, the addition/removal of investigation sites and the investigators, and/or the methods of investigation. These types of changing events need to be captured and linked with the timestamp they were captured on. These data (a type of time-series) are crucial for building time-series analysis of clinical entities and investigations. An example of time-series analysis may be determining the evolution of a vaccine over a certain time or between dates, or determining the role of certain investigators with a particular background in a clinical trial over a certain period linked with the trials’ stages or phases.
Processes
Data on-boarding: This is the process to identify and declare (if needed) the raw data schema Sch for a dataset D, identify and declare domain-specific terms (e.g., an ontology term), and declare limitations, risks, and use cases.
Data management: This is the process to bring the raw data under management that is already "on-boarded," as discussed above. It should declare and process data and type lineage [18] and be able to represent each data item with a timeline, hence treat every dataset as a time-series dataset. There is more on this in the following sections.
Data correlation: The framework is designed to on-board and manage the interrelated data coming from various resources. This process, which runs across all other processes, is meant to declare the correlations at each step to link technical terms to business/domain terms; for example, for a medical treatment for people with obesity, it should find all datasets that provide insights into these clinical terms. In each of the processes, this process is carried out at various levels with different semantics, as shown in the following sections.
Data analysis: This is the process to perform on-demand data processing based on pre-configured and orchestrated pipelines. When a scenario, Sce, is provided, the pipeline is triggered, and various orchestrated queries are processed by the framework generating derived results that can be streamed/sent to the user and additionally stored in the framework (particularly in the data lake) for future use.
Reproducibility: This is a process to reproduce [19] the result of a scenario in case of failure, doubt, or correctness checking of the framework system.
Architecture
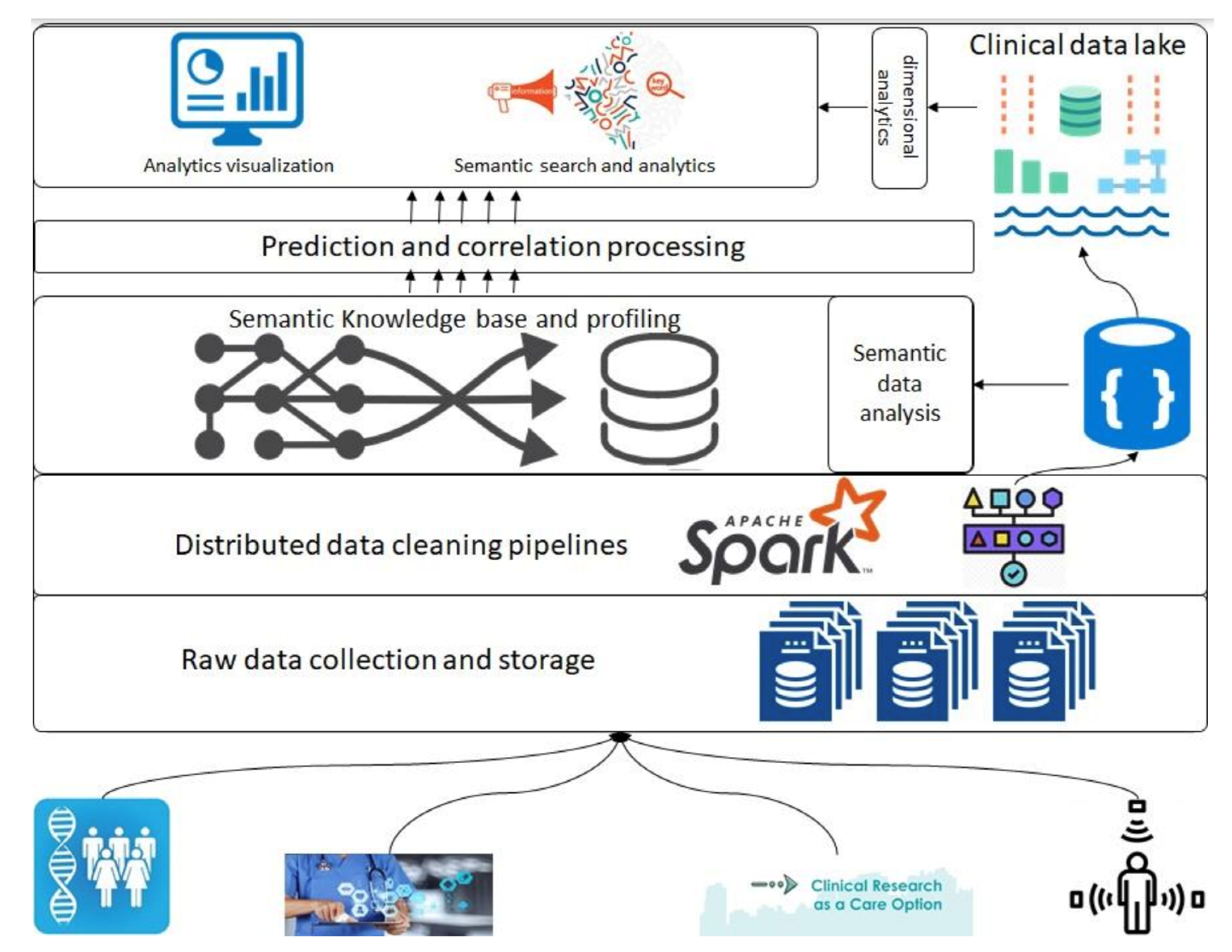
In this section, we introduce the overall architecture of the framework. Figure 1 presents an abstract overview of the generic architecture with different components that will address the above-described objectives. A detailed flow of the architecture follows in the following paragraphs; however, firstly, a layer-wise functioning is presented in the framework. This architecture consists of various components, such as raw data collection at the bottom that acquires data that can be regularly scraped from data sources at the bottom. Next is the data-cleaning pipeline powered by Apache Spark; it is the component that is based on defined schemas and transforms all datasets to a unified data format (such as Parquet) before storing them in the data lake. Next are the layers of semantic profiling and predictions. These layers are meant to process the data written to the data lake and perform analytical tasks such as fusing data from various sources (e.g., academic clinical articles, clinical trials, etc.) and predicting profiles from clinical trial data. Finally, the application layer at the top represents dashboards and external applications requesting data from the bottom layers through some services.
|
Before any dataset is brought into the system, the raw data schema and any vocabularies or ontologies must be declared. This initial step is called data “on-boarding,” and it will become more visible in the following section when processes are discussed. After this on-boarding process, the data flows through the layers discussed above as follows. Storage of raw data to the data lake (right top in Figure 1), performing quality checking and mappings declared in the on-boarding process, and storing to the clinical data lakehouse. The clinical data lake is also a file storage system with the ability to store each piece of data in a time-series format. For that purpose, we propose the usage of Apache Hudi [20], which allows each piece of data written to the data lake to be stamped/committed with a timestamp, thereby providing the ability to travel back in time. This data movement from the raw data to the data lake is optional in the sense that the stakeholders who want to store raw data can store it in the blob storage, whereas for others, the data can be directly brought to the data lake. In either case, the framework can stream the data either from an external source or from the raw data storage. For the streaming of the data, we propose the use of “Kafka” along with akka-streaming technologies such as “cloudflow.” [21]
Once the data are acquired and ingested into the data lake, queries can be performed to do analysis and perform data correlation between various data sources. The analysis and correlation can be carried out on both static and inflight data, i.e., data ingested in a streaming fashion, and hence requires the ability to merge/fuse with other streams.
References
Notes
This presentation is faithful to the original, with only a few minor changes to presentation, though grammar and word usage was substantially updated for improved readability. In some cases important information was missing from the references, and that information was added.