Journal:Fostering reproducibility, reusability, and technology transfer in health informatics
| Full article title | Fostering reproducibility, reusability, and technology transfer in health informatics |
|---|---|
| Journal | iScience |
| Author(s) | Hauschild, Anne-Christin; Eick, Lisa; Wienbeck, Joachim; Heider, Dominik |
| Author affiliation(s) | Philipps University of Marburg |
| Primary contact | Email: dominik dot heider at uni-marburg dot de |
| Year published | 2021 |
| Volume and issue | 24(7) |
| Article # | 102803 |
| DOI | 10.1016/j.isci.2021.102803 |
| ISSN | 2589-0042 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.sciencedirect.com/science/article/pii/S2589004221007719 |
| Download | https://www.sciencedirect.com/science/article/pii/S2589004221007719/pdfft (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
Computational methods can transform healthcare. In particular, health informatics combined with artificial intelligence (AI) has shown tremendous potential when applied in various fields of medical research and has opened a new era for precision medicine. The development of reusable biomedical software for research or clinical practice is time-consuming and requires rigorous compliance with quality requirements as defined by international standards.
However, research projects rarely implement such measures, hindering smooth technology transfer to the research community or manufacturers, as well as reproducibility and reusability.
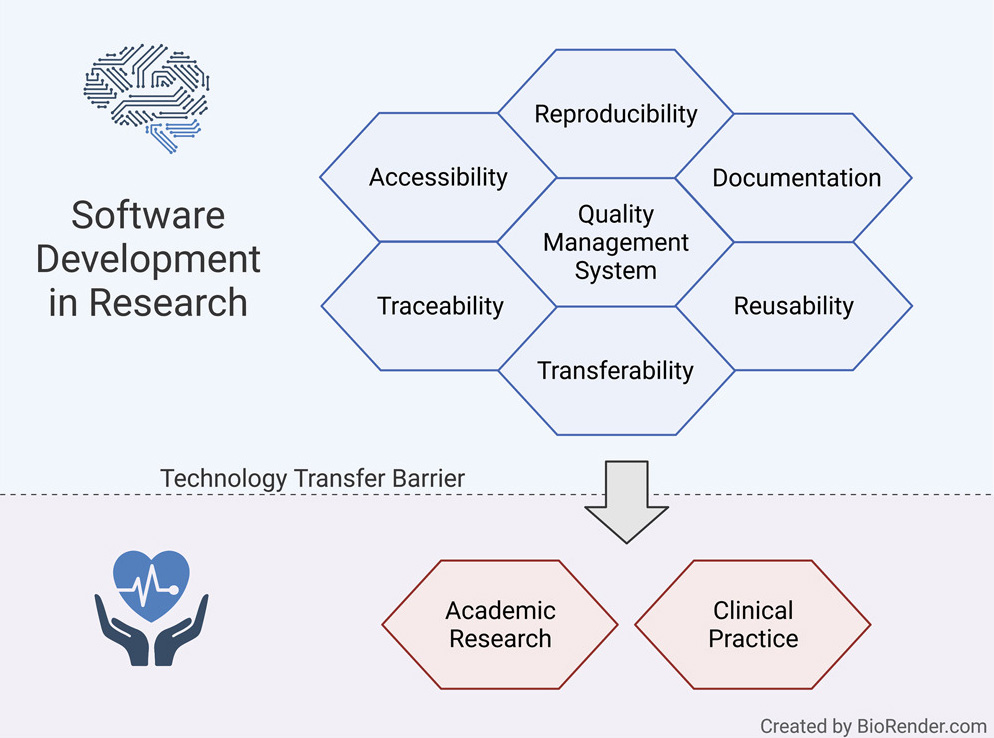
Here, we present a guideline for quality management systems (QMS) for academic organizations incorporating the essential components, while confining the requirements to an easily manageable effort. It provides a starting point to effortlessly implement a QMS tailored to specific needs and greatly facilitates technology transfer in a controlled manner, thereby supporting reproducibility and reusability.
Ultimately, the emerging standardized workflows can pave the way for an accelerated deployment in clinical practice.
Keywords: health informatics, bioinformatics, software engineering, software robustness
|
Introduction
Computational approaches offer new opportunities to transform health care. In particular, modern artificial intelligence (AI) and machine learning (ML) techniques have shown substantial potential when applied in various fields of medical research and therefore have opened up a new era for precision medicine. (Hawgood et al., 2015)[1] In the last decade, universities and other research organizations supported and encouraged by public funding agencies have allocated tremendous efforts to develop and enhance such predictive software models, algorithms, and “systems as medical devices” (SaMD) for clinical research and application.
A manifold of studies have proven AI to be advantageous for disease diagnosis, prognosis, and disease monitoring.[2][3] In cancer research, for instance, ML is used on omics data to gain deeper insights and understanding of the genetic and metabolic alterations that determine disease progression and enable tailored prognoses and monitoring.[4][5][6][7] Additionally, computational models on clinical information are used to assess individualized health risks, for instance, to identify high-risk patients for sepsis in intensive care units[8][9], the analysis of longitudinal data for the early detection of heart failure[10], or applications in infectious diseases.[11][12]
Once the outcome of this development is sufficiently mature and robust to be published and used in routine treatment or diagnosis, there is a need to transfer this knowledge to other research groups and, ultimately, clinical practice. Therefore, a straightforward technology transfer to a manufacturer of medical devices would be beneficial.[13][14]
Challenges of scientific software development for health
While the knowledge and technologies to develop effective and efficacious AI-driven clinical decision support systems exist, a manifold of pressing issues hinders further reuse in research and transfer to clinical practice. In software development projects in the industry, specialized developers typically work in large teams with considerable resources and are able to focus on usability and reusability.[15][16] In contrast, academic teams often consist of small groups of researchers (e.g., graduate students and postdoctoral scholars) who are typically not trained software engineers and only have temporary contracts and frequent turnover. Thus, individuals often develop software on a one-person-one-project basis.[13][16]
Moreover, funders and the academic hiring and promotion processes incentivize the pressure to publish and focus on “novelty” rather than software quality. Thus, it entices researchers to focus on theoretical aspects and proof of concept development.[13][17] Therefore, most researchers implement software in a prototype-centered manner, which often lacks quality checks such as systematic testing and may be published quickly.[18] However, these implementations often lack crucial qualities required for long-term reuse, such as documentation, usability, appropriate performance for real-life application, user-friendly interfaces, reusability, or minimized risk for potential users.[13][14]
Recently, scientific journals such as GigaScience or Biostatistics have promoted reproducibility and reusability by mandating FAIR principles (findability, accessibility, interoperability, and reusability). The concept of FAIR establishes a guideline for scientific data management and documentation.[13][19]
However, as recently surveyed by Pinto et al.[20], documentation of scientific software is one of the most significant “pain points.”[20] Journal publications are typically the primary source of documentation for scientific software and are quickly outdated by the agile software development style in academia.[16] Ideally, documentation should be detailed enough so that a developer with no prior knowledge of the project should make productive use of the software and use it for further development without biases or limitations.[13][18]
Another critical factor is accessibility. A lack of strict enforcement by journals, organizations, and funders has resulted in a loss of crucial data and software code.[13] According to an extensive analysis by Mangul et al., almost 28% of all resources linked in publications were not accessible, indicating poor maintenance. Moreover, a large proportion of software tested by Mangul et al. was not usable due to non-installability or lack of portability. The main inhibitors were storage locations outside of the journal’s directories or public versioning systems.[16][18]
Reproducibility and traceability are two of the most important aspects of biomedical and health informatics.[18][21] The lack of publicly available and comprehensible source code undermines the auditing of published methods and results. Additionally, the traceability of changes via version control is critical for reproducibility and reuse of research and code that replicators get to use.[18] These aspects ultimately undermine scientific rigor, transparency, and reproducibility.[13] The previously described accessibility, documentation, portability, and reusability factors are essential to ensure reproducibility and underpin trust in the scientific record of scientific software.[18]
Additionally, modern systems medicine approaches integrate all facets of private data such as electronic health records (EHR)[22], laboratory results[23], medical imaging[24], omics resources such as the cancer genome atlas (TCGA) or the gene expression omnibus[5][6][25][26], or pathway information.[4][6][7] However, sensitive patient data that enables an association of confidential personal information to single individuals underlies strict regulations such as the European General Data Protection Regulation (GDPR).[27] Therefore, the exchange within and among institutes is perceived as insurmountable, posing a roadblock hampering significant data-based medical innovations.
References
- ↑ Hawgood, Sam; Hook-Barnard, India G.; O’Brien, Theresa C.; Yamamoto, Keith R. (12 August 2015). "Precision medicine: Beyond the inflection point" (in en). Science Translational Medicine 7 (300): 300ps17–300ps17. doi:10.1126/scitranslmed.aaa9970. ISSN 1946-6234. https://stm.sciencemag.org/lookup/doi/10.1126/scitranslmed.aaa9970.
- ↑ Digital Health Center of Excellence (6 December 2017). "What are examples of Software as a Medical Device?". U.S. Food and Drug Administration. https://www.fda.gov/medical-devices/software-medical-device-samd/what-are-examples-software-medical-device.
- ↑ Fatima, Meherwar; Pasha, Maruf (2017). "Survey of Machine Learning Algorithms for Disease Diagnostic" (in en). Journal of Intelligent Learning Systems and Applications 09 (01): 1. doi:10.4236/jilsa.2017.91001. http://www.scirp.org/journal/PaperInformation.aspx?PaperID=73781&#abstract.
- ↑ 4.0 4.1 Batra, Richa; Alcaraz, Nicolas; Gitzhofer, Kevin; Pauling, Josch; Ditzel, Henrik J.; Hellmuth, Marc; Baumbach, Jan; List, Markus (1 December 2017). "On the performance of de novo pathway enrichment" (in en). npj Systems Biology and Applications 3 (1): 6. doi:10.1038/s41540-017-0007-2. ISSN 2056-7189. PMC PMC5445589. PMID 28649433. http://www.nature.com/articles/s41540-017-0007-2.
- ↑ 5.0 5.1 Hauschild, A.-C.; Baumbach, J.I.; Baumbach, J. (2012). "Integrated statistical learning of metabolic ion mobility spectrometry profiles for pulmonary disease identification". Genetics and Molecular Research 11 (3): 2733–2744. doi:10.4238/2012.July.10.17. http://www.funpecrp.com.br/gmr/year2012/vol11-3/pdf/gmr2065.pdf.
- ↑ 6.0 6.1 6.2 Jeanquartier, Fleur; Jean-Quartier, Claire; Kotlyar, Max; Tokar, Tomas; Hauschild, Anne-Christin; Jurisica, Igor; Holzinger, Andreas (2016), Holzinger, Andreas, ed., "Machine Learning for In Silico Modeling of Tumor Growth" (in en), Machine Learning for Health Informatics (Cham: Springer International Publishing) 9605: 415–434, doi:10.1007/978-3-319-50478-0_21, ISBN 978-3-319-50477-3, http://link.springer.com/10.1007/978-3-319-50478-0_21
- ↑ 7.0 7.1 Wiwie, Christian; Kuznetsova, Irina; Mostafa, Ahmed; Rauch, Alexander; Haakonsson, Anders; Barrio-Hernandez, Inigo; Blagoev, Blagoy; Mandrup, Susanne et al. (1 May 2019). "Time-Resolved Systems Medicine Reveals Viral Infection-Modulating Host Targets" (in en). Systems Medicine 2 (1): 1–9. doi:10.1089/sysm.2018.0013. ISSN 2573-3370. PMC PMC6524659. PMID 31119214. https://www.liebertpub.com/doi/10.1089/sysm.2018.0013.
- ↑ Calvert, Jacob; Saber, Nicholas; Hoffman, Jana; Das, Ritankar (13 February 2019). "Machine-Learning-Based Laboratory Developed Test for the Diagnosis of Sepsis in High-Risk Patients" (in en). Diagnostics 9 (1): 20. doi:10.3390/diagnostics9010020. ISSN 2075-4418. PMC PMC6468682. PMID 30781800. http://www.mdpi.com/2075-4418/9/1/20.
- ↑ Desautels, Thomas; Calvert, Jacob; Hoffman, Jana; Jay, Melissa; Kerem, Yaniv; Shieh, Lisa; Shimabukuro, David; Chettipally, Uli et al. (30 September 2016). "Prediction of Sepsis in the Intensive Care Unit With Minimal Electronic Health Record Data: A Machine Learning Approach" (in en). JMIR Medical Informatics 4 (3): e28. doi:10.2196/medinform.5909. ISSN 2291-9694. PMC PMC5065680. PMID 27694098. https://medinform.jmir.org/2016/3/e28/.
- ↑ Chen, Robert; Stewart, Walter F.; Sun, Jimeng; Ng, Kenney; Yan, Xiaowei (1 October 2019). "Recurrent Neural Networks for Early Detection of Heart Failure From Longitudinal Electronic Health Record Data: Implications for Temporal Modeling With Respect to Time Before Diagnosis, Data Density, Data Quantity, and Data Type" (in en). Circulation: Cardiovascular Quality and Outcomes 12 (10). doi:10.1161/CIRCOUTCOMES.118.005114. ISSN 1941-7713. PMC PMC6814386. PMID 31610714. https://www.ahajournals.org/doi/10.1161/CIRCOUTCOMES.118.005114.
- ↑ Heider, Dominik; Dybowski, Jan Nikolaj; Wilms, Christoph; Hoffmann, Daniel (1 December 2014). "A simple structure-based model for the prediction of HIV-1 co-receptor tropism" (in en). BioData Mining 7 (1): 14. doi:10.1186/1756-0381-7-14. ISSN 1756-0381. PMC PMC4124776. PMID 25120583. https://biodatamining.biomedcentral.com/articles/10.1186/1756-0381-7-14.
- ↑ Riemenschneider, Mona; Hummel, Thomas; Heider, Dominik (1 December 2016). "SHIVA - a web application for drug resistance and tropism testing in HIV" (in en). BMC Bioinformatics 17 (1): 314. doi:10.1186/s12859-016-1179-2. ISSN 1471-2105. PMC PMC4994198. PMID 27549230. http://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-016-1179-2.
- ↑ 13.0 13.1 13.2 13.3 13.4 13.5 13.6 13.7 Brito, Jaqueline J; Li, Jun; Moore, Jason H; Greene, Casey S; Nogoy, Nicole A; Garmire, Lana X; Mangul, Serghei (1 June 2020). "Recommendations to enhance rigor and reproducibility in biomedical research" (in en). GigaScience 9 (6): giaa056. doi:10.1093/gigascience/giaa056. ISSN 2047-217X. PMC PMC7263079. PMID 32479592. https://academic.oup.com/gigascience/article/doi/10.1093/gigascience/giaa056/5849489.
- ↑ 14.0 14.1 Riemenschneider, Mona; Wienbeck, Joachim; Scherag, André; Heider, Dominik (1 June 2018). "Data Science for Molecular Diagnostics Applications: From Academia to Clinic to Industry" (in en). Systems Medicine 1 (1): 13–17. doi:10.1089/sysm.2018.0002. ISSN 2573-3370. http://www.liebertpub.com/doi/10.1089/sysm.2018.0002.
- ↑ Guellec, D.; van Pottelsberghe de la Potterie, B. (14 June 2000). "The Impact of Public R&D Expenditure on Business R&D" (in en). OECD Science, Technology and Industry Working Papers 2000/04. doi:10.1787/670385851815. https://www.oecd-ilibrary.org/science-and-technology/the-impact-of-public-r-d-expenditure-on-business-r-d_670385851815.
- ↑ 16.0 16.1 16.2 16.3 Mangul, Serghei; Mosqueiro, Thiago; Abdill, Richard J.; Duong, Dat; Mitchell, Keith; Sarwal, Varuni; Hill, Brian; Brito, Jaqueline et al. (20 June 2019). "Challenges and recommendations to improve the installability and archival stability of omics computational tools" (in en). PLOS Biology 17 (6): e3000333. doi:10.1371/journal.pbio.3000333. ISSN 1545-7885. PMC PMC6605654. PMID 31220077. https://dx.plos.org/10.1371/journal.pbio.3000333.
- ↑ Mangul, Serghei; Martin, Lana S.; Eskin, Eleazar; Blekhman, Ran (1 December 2019). "Improving the usability and archival stability of bioinformatics software" (in en). Genome Biology 20 (1): 47, s13059–019–1649-8. doi:10.1186/s13059-019-1649-8. ISSN 1474-760X. PMC PMC6391762. PMID 30813962. https://genomebiology.biomedcentral.com/articles/10.1186/s13059-019-1649-8.
- ↑ 18.0 18.1 18.2 18.3 18.4 18.5 Lee, Graham; Bacon, Sebastian; Bush, Ian; Fortunato, Laura; Gavaghan, David; Lestang, Thibault; Morton, Caroline; Robinson, Martin et al. (1 February 2021). "Barely sufficient practices in scientific computing" (in en). Patterns 2 (2): 100206. doi:10.1016/j.patter.2021.100206. PMC PMC7892476. PMID 33659915. https://linkinghub.elsevier.com/retrieve/pii/S2666389921000167.
- ↑ Wilkinson, Mark D.; Dumontier, Michel; Aalbersberg, IJsbrand Jan; Appleton, Gabrielle; Axton, Myles; Baak, Arie; Blomberg, Niklas; Boiten, Jan-Willem et al. (1 December 2016). "The FAIR Guiding Principles for scientific data management and stewardship" (in en). Scientific Data 3 (1): 160018. doi:10.1038/sdata.2016.18. ISSN 2052-4463. PMC PMC4792175. PMID 26978244. http://www.nature.com/articles/sdata201618.
- ↑ 20.0 20.1 Pinto, Gustavo; Wiese, Igor; Dias, Luiz Felipe (1 March 2018). "How do scientists develop scientific software? An external replication". 2018 IEEE 25th International Conference on Software Analysis, Evolution and Reengineering (SANER) (Campobasso: IEEE): 582–591. doi:10.1109/SANER.2018.8330263. ISBN 978-1-5386-4969-5. http://ieeexplore.ieee.org/document/8330263/.
- ↑ Coiera, Enrico; Ammenwerth, Elske; Georgiou, Andrew; Magrabi, Farah (1 August 2018). "Does health informatics have a replication crisis?". Journal of the American Medical Informatics Association 25 (8): 963–968. doi:10.1093/jamia/ocy028. ISSN 1527-974X. PMC PMC6077781. PMID 29669066. https://doi.org/10.1093/jamia/ocy028.
- ↑ Shickel, Benjamin; Tighe, Patrick James; Bihorac, Azra; Rashidi, Parisa (1 September 2018). "Deep EHR: A Survey of Recent Advances in Deep Learning Techniques for Electronic Health Record (EHR) Analysis". IEEE Journal of Biomedical and Health Informatics 22 (5): 1589–1604. doi:10.1109/JBHI.2017.2767063. ISSN 2168-2194. PMC PMC6043423. PMID 29989977. https://ieeexplore.ieee.org/document/8086133/.
- ↑ Goecks, Jeremy; Jalili, Vahid; Heiser, Laura M.; Gray, Joe W. (1 April 2020). "How Machine Learning Will Transform Biomedicine" (in en). Cell 181 (1): 92–101. doi:10.1016/j.cell.2020.03.022. PMC PMC7141410. PMID 32243801. https://linkinghub.elsevier.com/retrieve/pii/S0092867420302841.
- ↑ Anwar, Syed Muhammad; Majid, Muhammad; Qayyum, Adnan; Awais, Muhammad; Alnowami, Majdi; Khan, Muhammad Khurram (1 November 2018). "Medical Image Analysis using Convolutional Neural Networks: A Review" (in en). Journal of Medical Systems 42 (11): 226. doi:10.1007/s10916-018-1088-1. ISSN 0148-5598. http://link.springer.com/10.1007/s10916-018-1088-1.
- ↑ Clough, Emily; Barrett, Tanya (2016), Mathé, Ewy; Davis, Sean, eds., "The Gene Expression Omnibus Database", Statistical Genomics (New York, NY: Springer New York) 1418: 93–110, doi:10.1007/978-1-4939-3578-9_5, ISBN 978-1-4939-3576-5, PMC PMC4944384, PMID 27008011, http://link.springer.com/10.1007/978-1-4939-3578-9_5
- ↑ Tomczak, Katarzyna; Czerwińska, Patrycja; Wiznerowicz, Maciej (2015). "The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge". Współczesna Onkologia 1A: 68–77. doi:10.5114/wo.2014.47136. ISSN 1428-2526. PMC PMC4322527. PMID 25691825. http://www.termedia.pl/doi/10.5114/wo.2014.47136.
- ↑ Voigt, Paul; von dem Bussche, Axel (2017) (in en). The EU General Data Protection Regulation (GDPR). Cham: Springer International Publishing. doi:10.1007/978-3-319-57959-7. ISBN 978-3-319-57958-0. http://link.springer.com/10.1007/978-3-319-57959-7.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation, though grammar and word usage was substantially updated for improved readability. In some cases important information was missing from the references, and that information was added. The original article lists references alphabetically, but this version—by design—lists them in order of appearance.