Difference between revisions of "Journal:From the desktop to the grid: Scalable bioinformatics via workflow conversion"
Shawndouglas (talk | contribs) (Added content. Saving and adding more.) |
Shawndouglas (talk | contribs) (Added content. Saving and adding more.) |
||
| Line 47: | Line 47: | ||
===Workflow engines=== | ===Workflow engines=== | ||
[[Galaxy (biomedical software)|Galaxy]]<ref name="BlankenbergGalaxy10" /> is a free web-based workflow system with several pre-installed tools for data-intensive biomedical research. Inclusion of arbitrary tools is reduced to the trivial task of creating ToolConfig<ref name="GalaxyTC">{{cite web |url=https://wiki.galaxyproject.org/Admin/Tools/ToolConfigSyntax |title=Admin/Tools/ToolConfigSyntax |publisher=Galaxy Project |date=19 June 2015 |accessdate=28 July 2015}}</ref> files, which are Extensible Markup Language documents (XML). The Galaxy project also features a so-called toolshed<ref name=" | [[Galaxy (biomedical software)|Galaxy]]<ref name="BlankenbergGalaxy10" /> is a free web-based workflow system with several pre-installed tools for data-intensive biomedical research. Inclusion of arbitrary tools is reduced to the trivial task of creating ToolConfig<ref name="GalaxyTC">{{cite web |url=https://wiki.galaxyproject.org/Admin/Tools/ToolConfigSyntax |title=Admin/Tools/ToolConfigSyntax |publisher=Galaxy Project |date=19 June 2015 |accessdate=28 July 2015}}</ref> files, which are Extensible Markup Language documents (XML). The Galaxy project also features a so-called toolshed<ref name="GalaxyToolshed">{{cite web |url=https://toolshed.g2.bx.psu.edu/ |title=Galaxy Tool Shed |publisher=Center for Comparative Genomics and Bioinformatics - Penn State |accessdate=07 July 2015}}</ref>, from which tools can be obtained and installed on Galaxy instances. At the time of writing Galaxy’s toolshed featured 3470 tools. However, we have found that Galaxy lacks extended support for popular workload managers and middlewares. | ||
Taverna<ref name="MissierTav10" /> offers an open-source and domain-independent suite of tools used to design and execute scientific workflows, helping users to automate and pipeline processing of data coming from different web services. At the time of writing Taverna features more than 3500 services available on startup and it also provides access to local and remote tools. Taverna allows users to track results and data flows with great granularity, since it implements the Open Provenance Model standard (OPM).<ref name="MoreauTheOpen11">{{cite journal |title=The Open Provenance Model core specification (v1.1) |journal=Future Generation Computer Systems |author=Moreau, L.; Clifford, B.; Freire, J. et al. |volume=27 |issue=6 |pages=743–756 |year=2011 |doi=10.1016/j.future.2010.07.005}}</ref> A very attractive feature of Taverna is the ability to share workflows via the [[myExperiment]] research environment.<ref name="">{{cite journal |title=myExperiment: a repository and social network for the sharing of bioinformatics workflows |journal=Nucleic Acids Research |author=Goble, C.; Bhagat, J.; Aleksejevs, S. et al. |volume=38 |issue=Supplemental 2 |pages=W677–W682 |year=May 2010 |doi=10.1093/nar/gkq429}}</ref> | Taverna<ref name="MissierTav10" /> offers an open-source and domain-independent suite of tools used to design and execute scientific workflows, helping users to automate and pipeline processing of data coming from different web services. At the time of writing Taverna features more than 3500 services available on startup and it also provides access to local and remote tools. Taverna allows users to track results and data flows with great granularity, since it implements the Open Provenance Model standard (OPM).<ref name="MoreauTheOpen11">{{cite journal |title=The Open Provenance Model core specification (v1.1) |journal=Future Generation Computer Systems |author=Moreau, L.; Clifford, B.; Freire, J. et al. |volume=27 |issue=6 |pages=743–756 |year=2011 |doi=10.1016/j.future.2010.07.005}}</ref> A very attractive feature of Taverna is the ability to share workflows via the [[myExperiment]] research environment.<ref name="">{{cite journal |title=myExperiment: a repository and social network for the sharing of bioinformatics workflows |journal=Nucleic Acids Research |author=Goble, C.; Bhagat, J.; Aleksejevs, S. et al. |volume=38 |issue=Supplemental 2 |pages=W677–W682 |year=May 2010 |doi=10.1093/nar/gkq429}}</ref> | ||
| Line 54: | Line 54: | ||
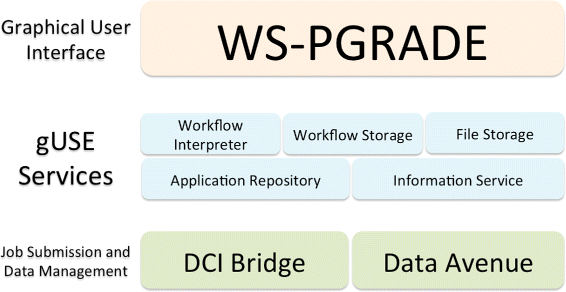
The grid and cloud User Support Environment (gUSE) offers an open-source, free, web-based workflow platform able to tap into distributed HPC infrastructures.<ref name="KacsukWS12" /> gUSE entails a set of components and services that offers access to distributed computing interfaces (DCI). The Web Services Parallel Grid Runtime and Developer Environment Portal (WS-PGRADE) component acts as the graphical user interface. This web-based portal is a series of dynamically generated web pages, through which users can create, execute, and monitor workflows. WS-PGRADE communicates with internal gUSE services (e.g., Workflow Interpreter, Workflow Storage, Information Service) using the Web Services Description Language (WSDL).<ref name="ChristensenWeb01">{{cite web |url=https://www.w3.org/TR/wsdl |title=Web Services Description Language (WSDL) 1.1 |author=Christensen, E.; Curbera, F.; Meredith, G.; Weerawarana, S. |publisher=World Wide Web Consortium |date=15 March 2001 |06 July 2016}}</ref> Passing documents in the WSDL format between its components allows gUSE services to interact with other workflow systems. Figure 1 shows the three-tiered architecture of gUSE. This complex and granular architecture of gUSE enables administrators to distribute the installation of gUSE across resources. A typical set-up is to install WS-PGRADE on a dedicated web server, while installing other services and components on more powerful computers. | The grid and cloud User Support Environment (gUSE) offers an open-source, free, web-based workflow platform able to tap into distributed HPC infrastructures.<ref name="KacsukWS12" /> gUSE entails a set of components and services that offers access to distributed computing interfaces (DCI). The Web Services Parallel Grid Runtime and Developer Environment Portal (WS-PGRADE) component acts as the graphical user interface. This web-based portal is a series of dynamically generated web pages, through which users can create, execute, and monitor workflows. WS-PGRADE communicates with internal gUSE services (e.g., Workflow Interpreter, Workflow Storage, Information Service) using the Web Services Description Language (WSDL).<ref name="ChristensenWeb01">{{cite web |url=https://www.w3.org/TR/wsdl |title=Web Services Description Language (WSDL) 1.1 |author=Christensen, E.; Curbera, F.; Meredith, G.; Weerawarana, S. |publisher=World Wide Web Consortium |date=15 March 2001 |06 July 2016}}</ref> Passing documents in the WSDL format between its components allows gUSE services to interact with other workflow systems. Figure 1 shows the three-tiered architecture of gUSE. This complex and granular architecture of gUSE enables administrators to distribute the installation of gUSE across resources. A typical set-up is to install WS-PGRADE on a dedicated web server, while installing other services and components on more powerful computers. | ||
[[File:Fig1 Garza BMCBioinformatics2016 17.gif|566px]] | |||
{{clear}} | |||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| border="0" cellpadding="5" cellspacing="0" width="566px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 1.''' The three-thiered gUSE’s architecture. The three tiers of gUSE’s architecture: WS-PGRADE acts as the user interface, the service layer handles e.g., file, workflow storage. The Job Submission and Data Management layer contains the DCI Bridge, which is responsible to access DCIs. Figure based on Gottdank's "gUSE in a Nutshell".<ref name="Gottdank_gUSE13">{{cite web |url=http://sourceforge.net/projects/guse/files/gUSE_in_a_Nutshell.pdf/download |format=PDF |title=gUSE in a Nutshell |author=Gottdank, T. |publisher=MTA SZTAKI Laboratory of Parallel and Distributed Systems |date=2013}}</ref></blockquote> | |||
|- | |||
|} | |||
|} | |||
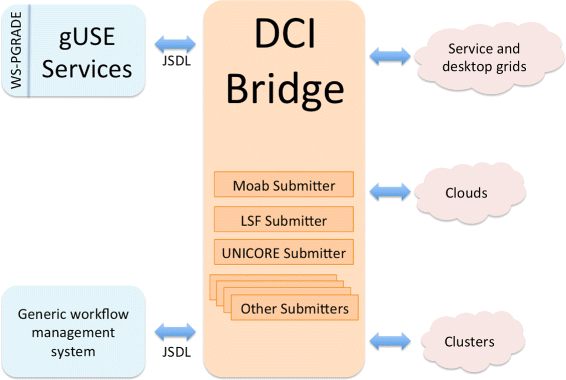
In order to provide a common workflow submission Application Programming Interface (API), gUSE channels workflow-related requests (i.e., start, monitor, cancel a job on a DCI) through the DCI Bridge component.<ref name="DCI15">{{cite web |url=http://sourceforge.net/projects/guse/files/3.7.1/Documentation/DCI_BRIDGE_MANUAL_v3.7.1.pdf/download |format=PDF |title=DCI Bridge Administrator Manual - Version 3.7.1 |publisher=MTA SZTAKI Laboratory of Parallel and Distributed Systems |date=12 June 2015}}</ref> The DCI Bridge is fully compatible with the Job Submission Description Language (JSDL)<ref name="AnjomshoaaJob05">{{cite web |url=https://www.ogf.org/documents/GFD.56.pdf |format=PDF |title=Job Submission Description Language (JSDL) Specification, Version 1.0 |author=Anjomshoaa, A.; Brisard, F.; Drescher, M. et al. |publisher=Global Grid Forum |date=07 November 2005}}</ref>, thus enabling other workflow management systems to interact with it in order to benefit from gUSE’s flexibility and DCI support. The DCI Bridge contains so-called DCI Submitters, each containing specific code to submit, monitor, cancel jobs on each of the supported DCIs (e.g., UNICORE<ref name="RombergTheUNI02">{{cite journal |title=The UNICORE Grid Infrastructure |journal=Scientific Programming |author=Romberg, M. |volume=10 |issue=2 |pages=149–157 |year=2002 |doi=10.1155/2002/483253}}</ref>, LSF<ref name="IBMSpec">{{cite web |url=http://www-03.ibm.com/systems/spectrum-computing/products/lsf/index.html |title=IBM Spectrum LSF |publisher=IBM Corporation |date=2012}}</ref>, Moab<ref name="ACProds">{{cite web |url=http://www.adaptivecomputing.com/products/hpc-products/ |title=HPC Products |publisher=Adaptive Computing |accessdate=06 July 2015}}</ref>). Figure 2 presents a schematic overview of the interaction between the DCI Bridge and other components. | |||
[[File:Fig2 Garza BMCBioinformatics2016 17.gif|566px]] | |||
{{clear}} | |||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| border="0" cellpadding="5" cellspacing="0" width="566px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 2.''' Schematic overview of gUSE’s DCI Bridge. Interaction of the DCI Bridge with gUSE services and other workflow management systems is done via JSDL requests. The DCI Bridge contains DCI Submitters, which contain specific code for each of the supported DCIs in gUSE. Figure based on MTA SZTAKI Laboratory of Parallel and Distributed Systems' ''DCI Bridge Administrator Manual''.<ref name="DCI15" /></blockquote> | |||
|- | |||
|} | |||
|} | |||
==References== | ==References== | ||
| Line 59: | Line 85: | ||
==Notes== | ==Notes== | ||
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. The original citation #1 was a mix of two different sources, and it has been corrected here to refer in full to the correct citation from ''The Economist''. | This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. The original citation #1 was a mix of two different sources, and it has been corrected here to refer in full to the correct citation from ''The Economist''. The original ctation #47 got bumped up to 19 due to mandatory wiki ordering, bumping other original citation numbers down one. | ||
<!--Place all category tags here--> | <!--Place all category tags here--> | ||
Revision as of 20:53, 13 June 2016
| Full article title | From the desktop to the grid: Scalable bioinformatics via workflow conversion |
|---|---|
| Journal | BMC Bioinformatics |
| Author(s) | de la Garza, Luis; Veit, Johannes; Szolek, Andras; Röttig, Marc; Aiche, Stephan; Gesing, Sandra; Reinert, Knut; Kohlbacher, Oliver |
| Author affiliation(s) | University of Tübingen, Freie Universität Berlin, University of Notre Dame |
| Primary contact | Email: delagarza [at] informatik [dot] uni-tuebingen [dot] de |
| Year published | 2016 |
| Volume and issue | 17 |
| Page(s) | 127 |
| DOI | 10.1186/s12859-016-0978-9 |
| ISSN | 1471-2105 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-016-0978-9 |
| Download | https://bmcbioinformatics.biomedcentral.com/track/pdf/10.1186/s12859-016-0978-9 (PDF) |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
Background
Reproducibility is one of the tenets of the scientific method. Scientific experiments often comprise complex data flows, selection of adequate parameters, and analysis and visualization of intermediate and end results. Breaking down the complexity of such experiments into the joint collaboration of small, repeatable, well defined tasks, each with well defined inputs, parameters, and outputs, offers the immediate benefit of identifying bottlenecks, pinpoint sections which could benefit from parallelization, among others. Workflows rest upon the notion of splitting complex work into the joint effort of several manageable tasks.
There are several engines that give users the ability to design and execute workflows. Each engine was created to address certain problems of a specific community, therefore each one has its advantages and shortcomings. Furthermore, not all features of all workflow engines are royalty-free — an aspect that could potentially drive away members of the scientific community.
Results
We have developed a set of tools that enables the scientific community to benefit from workflow interoperability. We developed a platform-free structured representation of parameters, inputs, outputs of command-line tools in so-called Common Tool Descriptor documents. We have also overcome the shortcomings and combined the features of two royalty-free workflow engines with a substantial user community: the Konstanz Information Miner, an engine which we see as a formidable workflow editor, and the Grid and User Support Environment, a web-based framework able to interact with several high-performance computing resources. We have thus created a free and highly accessible way to design workflows on a desktop computer and execute them on high-performance computing resources.
Conclusions
Our work will not only reduce time spent on designing scientific workflows, but also make executing workflows on remote high-performance computing resources more accessible to technically inexperienced users. We strongly believe that our efforts not only decrease the turnaround time to obtain scientific results but also have a positive impact on reproducibility, thus elevating the quality of obtained scientific results.
Keywords
Workflow, interoperability, KNIME, grid, cloud, Galaxy, gUSE
Background
The importance of reproducibility for the scientific community has been a topic lately discussed in both high-impact scientific publications and popular news outlets.[1][2] To be able to independently replicate results — be it for verification purposes or to further advance research — is important for the scientific community. Therefore, it is crucial to structure an experiment in such a way that reproducibility could be easily achieved.
Workflows are structured, abstract recipes that help users construct a series of steps in an organized way. Each step is a parametrised specific action that receives some input and produces some output. The collective execution of these steps is seen as a domain-specific task.
With the availability of biological big data, the need to represent workflows in computing languages has also increased.[3] Scientific tasks such as genome comparison, mass spectrometry analysis, protein-protein interaction, just to name a few, access extensive datasets. Currently, a vast number of workflow engines exist[4][5][6][7][8], and each of these technologies has amassed a considerable user base. These engines support, in some way or another, the execution of workflows on distributed high-performance computing (HPC) resources (e.g., grids, clusters, clouds, etc.), thus allowing speedier obtention of results. A wise selection of a workflow engine will shorten the time spent between workflow design and retrieval of results.
Workflow engines
Galaxy[6] is a free web-based workflow system with several pre-installed tools for data-intensive biomedical research. Inclusion of arbitrary tools is reduced to the trivial task of creating ToolConfig[9] files, which are Extensible Markup Language documents (XML). The Galaxy project also features a so-called toolshed[10], from which tools can be obtained and installed on Galaxy instances. At the time of writing Galaxy’s toolshed featured 3470 tools. However, we have found that Galaxy lacks extended support for popular workload managers and middlewares.
Taverna[7] offers an open-source and domain-independent suite of tools used to design and execute scientific workflows, helping users to automate and pipeline processing of data coming from different web services. At the time of writing Taverna features more than 3500 services available on startup and it also provides access to local and remote tools. Taverna allows users to track results and data flows with great granularity, since it implements the Open Provenance Model standard (OPM).[11] A very attractive feature of Taverna is the ability to share workflows via the myExperiment research environment.[12]
The Konstanz Information Miner Analytics Platform (KNIME Analytics Platform)[4][13] is a royalty-free engine that allows users to build and execute workflows using a powerful and user-friendly interface. The KNIME Analytics Platform comes preloaded with several ready-to-use tasks (called KNIME nodes) that serve as the building stones of a workflow. It is also possible to extend the KNIME Analytics Platform by either downloading community nodes or building custom nodes using a well-documented process.[14][15] Workflows executed on the KNIME Analytics Platform are limited to run on the same personal computer on which it has been installed, thus rendering it unsuitable for tasks with high-memory or high-performance requirements. KNIME is offered in two variants able to execute workflows on distributed HPC resources: KNIME Cluster Execution[16], and KNIME Server.[17] These two suites are, however, royalty-based — an aspect that might shy away users of the scientific community.
The grid and cloud User Support Environment (gUSE) offers an open-source, free, web-based workflow platform able to tap into distributed HPC infrastructures.[5] gUSE entails a set of components and services that offers access to distributed computing interfaces (DCI). The Web Services Parallel Grid Runtime and Developer Environment Portal (WS-PGRADE) component acts as the graphical user interface. This web-based portal is a series of dynamically generated web pages, through which users can create, execute, and monitor workflows. WS-PGRADE communicates with internal gUSE services (e.g., Workflow Interpreter, Workflow Storage, Information Service) using the Web Services Description Language (WSDL).[18] Passing documents in the WSDL format between its components allows gUSE services to interact with other workflow systems. Figure 1 shows the three-tiered architecture of gUSE. This complex and granular architecture of gUSE enables administrators to distribute the installation of gUSE across resources. A typical set-up is to install WS-PGRADE on a dedicated web server, while installing other services and components on more powerful computers.
|
In order to provide a common workflow submission Application Programming Interface (API), gUSE channels workflow-related requests (i.e., start, monitor, cancel a job on a DCI) through the DCI Bridge component.[20] The DCI Bridge is fully compatible with the Job Submission Description Language (JSDL)[21], thus enabling other workflow management systems to interact with it in order to benefit from gUSE’s flexibility and DCI support. The DCI Bridge contains so-called DCI Submitters, each containing specific code to submit, monitor, cancel jobs on each of the supported DCIs (e.g., UNICORE[22], LSF[23], Moab[24]). Figure 2 presents a schematic overview of the interaction between the DCI Bridge and other components.
|
References
- ↑ "Unreliable research: Trouble at the lab". The Economist 409 (8858): 26–30. 19 October 2013. http://www.economist.com/news/briefing/21588057-scientists-think-science-self-correcting-alarming-degree-it-not-trouble. Retrieved 07 July 2015.
- ↑ McNutt, M. (17 January 2014). "Reproducibility". Science 343 (6168): 229. doi:10.1126/science.1250475. PMID 24436391.
- ↑ Greene, C.S.; Tan, J.; Ung, M.; Moore, J.H.; Cheng, C. (2014). "Big data bioinformatics". Journal of Cellular Physiology 229 (12): 1896-900. doi:10.1002/jcp.24662. PMID 24799088.
- ↑ 4.0 4.1 Berthold, M.R.; Cebron, N.; Dill, F.; Gabriel, T.R.; Kötter, T.; Meinl, T.; Ohl, P.; Sieb, C.; Thiel, K.; Wiswedel, B. (2008). "Chapter 38: KNIME: The Konstanz Information Miner". In Preisach, C.; Burkhardt, H.; Schmidt-Thieme, L.; Decker, R.. Data Analysis, Machine Learning and Applications. Springer Berlin Heidelberg. doi:10.1007/978-3-540-78246-9_38. ISBN 9783540782391.
- ↑ 5.0 5.1 Kacsuk, P.; Farkas, Z.; Kozlovszky, M.; Hermann, G.; Balasko, A.; Karoczkai, K.; Marton, I. (2012). "WS-PGRADE/gUSE Generic DCI Gateway Framework for a Large Variety of User Communities". Journal of Grid Computing 10 (4): 601–630. doi:10.1007/s10723-012-9240-5.
- ↑ 6.0 6.1 Blankenberg, D.; Von Kuster, G.; Coraor, N.; Ananda, G.; Lazarus, R.; Mangan, M.; Nekrutenko, A.; Taylor, J. (2010). "Galaxy: A web-based genome analysis tool for experimentalists". Current Protocols in Molecular Biology 19 (Unit 19.10.1–21). doi:10.1002/0471142727.mb1910s89. PMC PMC4264107. PMID 20069535. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4264107.
- ↑ 7.0 7.1 Missier, P.; Soiland-Reyes, S.; Owen, S.; Tan, W.; Nenadic, A.; Dunlop, I.; Williams, A.; Oinn, T.; Goble, C. (2010). "Chapter 33: Taverna, Reloaded". In Gertz, M.; Ludäscher, B.. Data Analysis, Machine Learning and Applications. Springer Berlin Heidelberg. doi:10.1007/978-3-642-13818-8_33. ISBN 9783642138171.
- ↑ Abouelhoda, M.; Issa, S.A.; Ghanem, M. (2012). "Tavaxy: Integrating Taverna and Galaxy workflows with cloud computing support". BMC Bioinformatics 13: 77. doi:10.1186/1471-2105-13-77. PMC PMC3583125. PMID 22559942. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3583125.
- ↑ "Admin/Tools/ToolConfigSyntax". Galaxy Project. 19 June 2015. https://wiki.galaxyproject.org/Admin/Tools/ToolConfigSyntax. Retrieved 28 July 2015.
- ↑ "Galaxy Tool Shed". Center for Comparative Genomics and Bioinformatics - Penn State. https://toolshed.g2.bx.psu.edu/. Retrieved 07 July 2015.
- ↑ Moreau, L.; Clifford, B.; Freire, J. et al. (2011). "The Open Provenance Model core specification (v1.1)". Future Generation Computer Systems 27 (6): 743–756. doi:10.1016/j.future.2010.07.005.
- ↑ Goble, C.; Bhagat, J.; Aleksejevs, S. et al. (May 2010). "myExperiment: a repository and social network for the sharing of bioinformatics workflows". Nucleic Acids Research 38 (Supplemental 2): W677–W682. doi:10.1093/nar/gkq429.
- ↑ "KNIME: Open for Innovation". KNIME.org AG. http://www.knime.org/. Retrieved 29 June 2015.
- ↑ "New Node Wizard". Developer Guide. KNIME.org AG. https://tech.knime.org/new-node-wizard. Retrieved 06 July 2015.
- ↑ "Community Contributions". KNIME Community. KNIME.org AG. https://tech.knime.org/community. Retrieved 07 July 2015.
- ↑ "KNIME Cluster Execution". Products. KNIME.org AG. https://www.knime.org/cluster-execution. Retrieved 06 July 2015.
- ↑ "KNIME Server - The Heart of a Collaborative KNIME Setup". Products. KNIME.org AG. https://www.knime.org/knime-server. Retrieved 06 July 2015.
- ↑ Christensen, E.; Curbera, F.; Meredith, G.; Weerawarana, S. (15 March 2001). "Web Services Description Language (WSDL) 1.1". World Wide Web Consortium. https://www.w3.org/TR/wsdl.
- ↑ Gottdank, T. (2013). "gUSE in a Nutshell" (PDF). MTA SZTAKI Laboratory of Parallel and Distributed Systems. http://sourceforge.net/projects/guse/files/gUSE_in_a_Nutshell.pdf/download.
- ↑ 20.0 20.1 "DCI Bridge Administrator Manual - Version 3.7.1" (PDF). MTA SZTAKI Laboratory of Parallel and Distributed Systems. 12 June 2015. http://sourceforge.net/projects/guse/files/3.7.1/Documentation/DCI_BRIDGE_MANUAL_v3.7.1.pdf/download.
- ↑ Anjomshoaa, A.; Brisard, F.; Drescher, M. et al. (7 November 2005). "Job Submission Description Language (JSDL) Specification, Version 1.0" (PDF). Global Grid Forum. https://www.ogf.org/documents/GFD.56.pdf.
- ↑ Romberg, M. (2002). "The UNICORE Grid Infrastructure". Scientific Programming 10 (2): 149–157. doi:10.1155/2002/483253.
- ↑ "IBM Spectrum LSF". IBM Corporation. 2012. http://www-03.ibm.com/systems/spectrum-computing/products/lsf/index.html.
- ↑ "HPC Products". Adaptive Computing. http://www.adaptivecomputing.com/products/hpc-products/. Retrieved 06 July 2015.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. The original citation #1 was a mix of two different sources, and it has been corrected here to refer in full to the correct citation from The Economist. The original ctation #47 got bumped up to 19 due to mandatory wiki ordering, bumping other original citation numbers down one.