Journal:Generating big data sets from knowledge-based decision support systems to pursue value-based healthcare
| Full article title | Generating big data sets from knowledge-based decision support systems to pursue value-based healthcare |
|---|---|
| Journal | International Journal of Interactive Multimedia and Artificial Intelligence |
| Author(s) | González-Ferrer, Arturo; Seara, Germán; Cháfer, Joan; Mayol, Julio |
| Author affiliation(s) | Instituto de Investigación Sanitaria San Carlos, Hospital Universitario Clínico San Carlos |
| Primary contact | Email: arturogf at gmail dot com |
| Year published | 2018 |
| Volume and issue | 4 (7) |
| Page(s) | 42–46 |
| DOI | 10.9781/ijimai.2017.03.006 |
| ISSN | 1989-1660 |
| Distribution license | Creative Commons Attribution 3.0 Unported |
| Website | http://www.ijimai.org/journal/node/1626 |
| Download | http://www.ijimai.org/journal/sites/default/files/files/2017/03/ijimai_4_7_6_pdf_12585.pdf (PDF) |
Abstract
Talking about big data in healthcare we usually refer to how to use data collected from current electronic medical records, either structured or unstructured, to answer clinically relevant questions. This operation is typically carried out by means of analytics tools (e.g., machine learning) or by extracting relevant data from patient summaries through natural language processing techniques. From other perspectives of research in medical informatics, powerful initiatives have emerged to help physicians make decisions, in both diagnostics and therapeutics, built from existing medical evidence (i.e., knowledge-based decision support systems). Many of the problems these tools have shown, when used in real clinical settings, are related to their implementation and deployment, more than failing in their support; however, technology is slowly overcoming interoperability and integration issues. Beyond the point-of-care decision support these tools can provide, the data generated when using them, even in controlled trials, could be used to further analyze facts that are traditionally ignored in the current clinical practice. In this paper, we reflect on the technologies available to make the leap and how they could help drive healthcare organizations shifting to a value-based healthcare philosophy.
Keywords: big data, DSS, e-health, knowledge management, management systems
Introduction
Healthcare made a big step towards modernization with the emergence of the evidence-based medicine (EBM) concept in the late 1980s.[1] EBM is an approach to medical practice that aims to apply the best known scientific evidence into clinical decision-making regarding diagnosis and effective management of specific conditions and diseases. While the EBM concept was generally well received by care professionals, many factors — such as their daily work conditions or their high work load — affect putting into practice this approach in the expected way. A recent report from the Institute of Medicine in 2012 revealed that only 10 to 20 percent of the decisions clinicians make are evidence-based.[2] This fact reflects the need for medical practitioners, supported by their healthcare organizations, to make a shift in their behavior about the way clinical practice is currently carried out.
The idea of EBM emerged in very different conditions from the current scenario. An explosion of technical possibilities — in nearly thirty years — have come into place to help organizations take a more modern approach, providing them with support in this regard. Not only epidemiological research can drive EBM, but also new data-oriented approaches. When saying “data-oriented,” we refer to data about the real daily clinical practice: how, when, why, and by whom are clinical actions carried out (or not), and what are the health results of those actions. Nonetheless, this might still be hampered by the current design of electronic medical records (EMRs) and by the role and focus that contemporary doctors should adopt. The use of EMRs by physicians could be insufficient, as recognized by studies[2] that expose that, even after post-digitalization of healthcare, they are not utilized to their maximum potential at all.
The fact that the EBM approach was crafted with the goal in mind of pursuing effectiveness in disease management left behind the consideration of organizational and human factors that are crucial in how decisions are truly made. By analyzing data generated by healthcare organizations we could yield information concerning the pitfalls that are hindering evidence-based clinical actions. At the same time, new evidence could be unveiled that is probably not considered in the current production of clinical practice guidelines (CPGs). For example, Toussi et al.[3] used data mining techniques to find out how physicians prescribe medications in diverse cases with various clinical conditions, in order to complement existing clinical guidelines where absence of enough evidence occur. Furthermore, specific training actions could be directed to address common failures detected in the management of medical conditions.
Therefore, the problem that healthcare organizations are trying to solve, under the hypothesis that the “big data” paradigm will change the way clinical practice is currently carried out, is how they can produce data that help to unveil real clinical behavior and "mindlines,"[4] linked with other organizational data (e.g., costs) and context information that could be behind their actions and decisions. Only making this analysis possible will they be able to change their philosophy to pursue and underpin value, beyond so-called effectiveness. And value here means detecting which actions — later possibly abstracted into policies — could really improve the behavior of the organizations and care professionals for the better care of their patients.
In this paper, we intend to reflect on some existing techniques beyond current electronic medical records (EMRs) that can help to generate such data sets, as well as considerations to be made, providing some examples of initiatives we are trying to push forward from the Innovation Unit of Hospital Universitario Clínico San Carlos (HUCSC).
Knowledge-based decision support systems (KB-DSS)
In 2007, Gartner[5] reported a five-stage evolution model for electronic health records (EHRs) where they established a path of characteristics, in terms of eight core capabilities, that EHRs should follow in order to provide the proper support to care professionals. Systems complying with Generation 3 requisites were supposed to be able to bring evidence-based medicine to the point of care, and theoretically coincide with the capabilities of most available EHRs, which progressed mainly through the core capabilities of system management, interoperability, and clinical data models, though there is still space for improvement today. Generation 4 was expected to improve the core capabilities of decision support, clinically relevant data analysis, presentation, and clinical workflow management.
More recently, Greenes offered his view about the past and future of knowledge-driven health IT[6], stating that current EHR systems were built for a model that is now old and even inappropriate, supported by proprietary infrastructures and knowledge content. He also mentioned the gradual increase in knowledge-based applications during the 2000s, with the creation of computer-interpretable clinical guideline formalisms like GLIF[7] and others.[8] By that time, these systems were having little penetration into real clinical settings, mostly due to the lack of pervasiveness of standards and the use of proprietary tools. Fortunately, this fact is something widely recognized by the current health IT community, and steps have been taken to tackle these problems. From requirements analysis of data standards[9] and development data integration mechanisms[10] for making DSS interoperable, the emergence of new lightweight web services standards like the HL7 Fast Healthcare Interoperability Resources (FHIR)[11], to substantial investments from public bodies that ended up with real deployments and piloting of patient guidance systems. A good example is MobiGuide[10][12], a project funded by the European Commission under the seventh framework program (FP7). Its goal was to create an intelligent KB-DSS to help physicians and patients taking the most appropriate decisions to manage concrete conditions (atrial fibrillation, gestational diabetes) using a back-end server and wearable sensors to monitor patients' status.
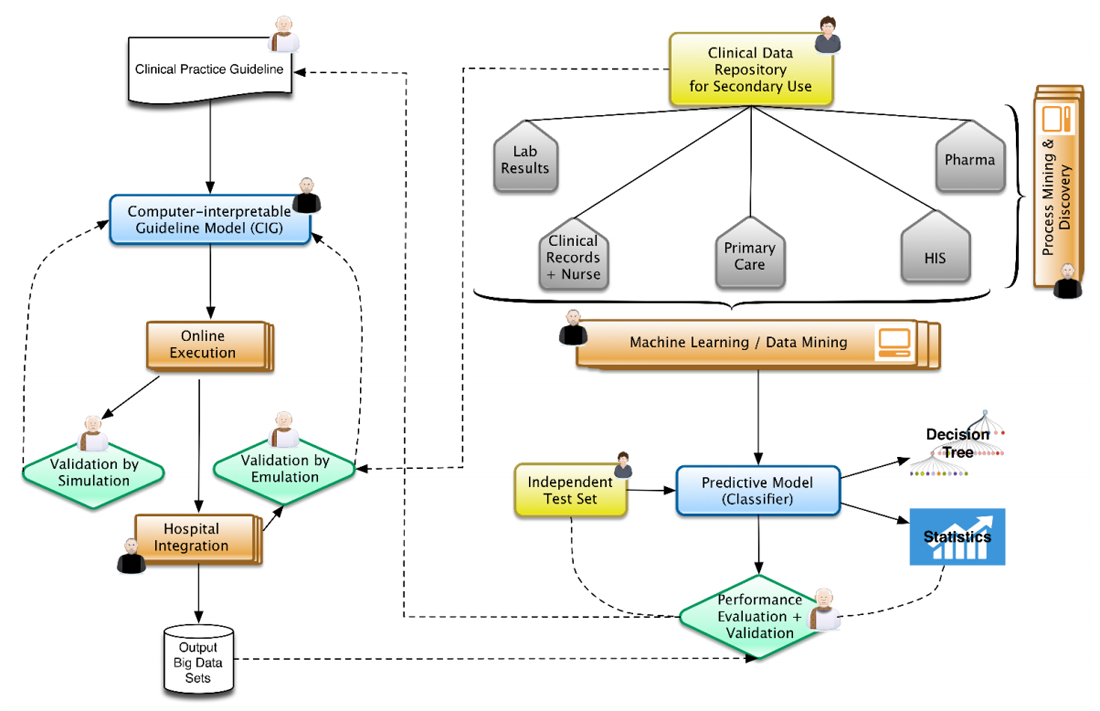
In this context, Figure 1 represents the architecture that represents our view, well aligned to positions already expressed by some research communities.[13] From top to bottom and left to right, physicians and epidemiologists develop CPGs that can be computerized, together with knowledge engineers, into computer-interpretable guideline (CIG) models. With the proper validation mechanisms, using data previously aggregated into clinical data repositories, these models can be trialed, after the corresponding integration into hospital information systems. The execution of CIG models can start generating data sets that are composed of acceptance or denials by physicians of recommendations (e.g., diagnosis, drug prescriptions, therapies, etc.) provided by the knowledge-based DSS developed, and treatments paths followed for different patient profiles. These paths can later on be analyzed by means of process mining techniques[14][15], unveiling common practices followed while using decision support and comparing the compliance of traditional clinical practice with the one recommended by the evidence-based DSS. At the same time, normalized clinical data repositories, while ensuring the quality of the data stored, can be used in the traditional view of machine learning and big data research.[16][17] The results could be complemented by comparing them with the output data sets of the KB-DSS. The output of the research could provide new evidence to be included in new versions of the CPGs (continuous improvement).
|
Innovative projects in HUCSC
The Innovation Unit of Hospital Universitario Clínico San Carlos, being transversal to the healthcare institution, is intended to cover two main aspects of innovation, always pursuing to increase value. On the one hand, it is expected to help hospital professionals to get their research into the mainstream, when there is an opportunity for it. On the other hand, it maintains a technical department to develop innovative products and test their prototypes, driving the hospital to maximize the possibilities that technological solutions could provide, especially artificial intelligence-based tools.
The ultimate intention is to disseminate the existence of these techniques while facilitating its understanding, create a culture of innovation within the hospital, and, when possible, get external companies to finalize these prototypes (or collaborate in the development) if they are demonstrated relevant and close to market possibility. The following are several ongoing projects aligned with these goals and that contribute several methods and artifacts to the architecture presented.
Computer-interpretable guideline for diagnosis and treatment of hyponatremia
The endocrinology department demanded a process-based solution to help new residents to improve their ability to diagnose and manage the hyponatremia condition (presenting low levels of serum sodium). Hyponatremia is the most frequent electrolyte disorder; however, according to some studies, it has proved to be very difficult to comprehend by physicians in general.[18] To address this project, we developed a CIG model[19][20] using the PROForma set of tools[21][22], covering the diagnosis of hyponatremia, classifying it into thirteen different subtypes. During a retrospective validation of the system with the data from 65 patients, we compared the system’s output to the diagnosis consensus of two experts, obtaining a very high agreement (kappa=0.86). The agreement found was also higher than a previous experiment found in the literature[23], carried out by comparing the performance of a resident physician — using the original paper guideline — with the diagnosis of senior physicians. Nonetheless, the most relevant advance of using such a system, beyond its successful diagnosis performance, was the identification and recording of data cases that were contrary to the consensus of international hyponatremia experts, specifically regarding hypoaldosteronism, where concrete marker thresholds were thought to be associated to its diagnosis. The application of our model found several cases where this hypothesis did not apply, showing the lack of real evidence and the need for further research. This is a concrete demonstration of how putting into practice these knowledge-based systems can help detect where evidence is failing and focus new research directions.
Unsupervised learning of discharge data (big data)
The syndrome of inappropriate antidiuretic hormone secretion (SIADH) represents around one-third of all cases of hyponatremia. We carried out a project[24] to identify clusters of hospitalized SIADH patients sharing diagnosed pathologies (comorbidities), where the results coincided and extended previous research identifying individual comorbidities.
Our methods included testing of two different distance measures and hierarchical agglomerative clustering. We used similarity profile analysis for determination of the number of significant clusters and membership of individuals[25] (by means of the SIMPROF method included in the clustsig R package). The method provides also the members of each proposed cluster, where validation of the clusters produced is assessed by iteratively carrying out hundreds of permutations tests. Analyzing the data from around 650 patients, it unveiled eight clusters, with five of them being significant: cancer patients, urinary tract infection patients, patients with renal failure, patients with respiratory problems, and patients with atrial fibrillation and other heart conditions.
We found one main problem: this process is costly to carry out on a personal computer, especially when having thousands of columns (variables) in the data. We are evaluating the use of the Cloudera big data framework along with Apache Mahout[26] to build the next stage of scalable algorithms that are able to cope with big data sets. If successful, this should be accompanied by the deployment of a private cloud infrastructure[27] able to provide a machine learning as a service (MLaaS) platform, due to the characteristics of patient sensitive data.
Hikari: A case study of mental health (big data)
In June 2015, Fujitsu Laboratories of Europe Ltd. and Fujitsu EMEIA in Spain signed a strategic research collaboration agreement with the Foundation for Biomedical Research of Hospital Clínico San Carlos (FIBHCSC). Mental health was selected as a key target for the initial project for several reasons: 1) the high levels of disability and morbidity associated to mental illness; 2) the important burden that mental illness imposes on patients, both at the individual and social level, and on the use of healthcare resources; and 3) the virtual impossibility to analyze results and its value, despite an apparently perfect design and theoretical structure of mental health services.[28]
Hikari, the Japanese word for light, is a part of Fujitsu’s Zinrai Artificial Intelligence technologies focused on people that includes data analytics and semantic modeling. In this project we have used relevant dissociated clinical data from the psychiatric department, obtained during the last 10 years, including patient discharge records and the specific registries of psychiatric emergency care, in order to generate a very simple and friendly tool that allows clinicians to have access to information related to the main diagnosis, comorbidities, and associated health risks, and also the possibility of analysis at the population level. It has been also useful to track the pathways through the healthcare system followed by patients, and to analyze the impact on the use of resources and costs.
At the present time, the database includes approximately 30,000 emergency care records and 6,500 hospitalizations; however, we expect that by the time this paper will be published, it will include data from more than 370,000 outpatients and 38,000 records of day hospital care. This will help us to establish patterns of behavior of the different pathologies and conditions in terms of comorbidities, pathways, and use of resources.
Clinical data repository for secondary use
Health observatories, regardless of regional, national, or supranational level, rely on reporting data that will inform on healthcare structure and compliance with programs or pathways. However, data on health outcomes and results are very few or close to none. This is very closely related to the incoherence and fragmented evolution of health care information systems.
In the last decades it has become increasingly evident the demographic and social change in Western societies that has brought the concepts of chronicity, fragility, and complexity of patients. This makes the continuity of care centered on patients an absolute necessity if we are to keep our health systems sustainable. Probably one of the main factors involved in this kind of transformation is access to daily care data that will enable patients, professionals, managers, and health policy makers to address these challenges.
If we consider the previous lines, it becomes more and more evident the desirability of having repositories of relevant dissociated clinical data that will allow the evaluation of the procedures and results of the real clinical practice, to compare them with recommendations based on evidence, and, at the same time, to generate new evidence from the stored data. It is essential to standardize data structure, context (actors, themes, time), continuity of care (such as UNE-EN-13940), generic reference models (such as UNE-EN-ISO 13606, part1), understandable archetypes for clinicians (such as UNE-EN-ISO 13606, part2), terminologies (such as SNOMED-CT), and ontologies for knowledge representation.[29] And, of course, it's vital to fulfill the criteria of privacy and data security provided in the legislation, recently renewed in Europe with a new regulation.[30]
Discussion
The application of KB-DSS in healthcare can provide diverse revelations. One of the most useful can be the detection of mistakes incurred frequently by professionals when comparing to evidence-based guidelines. Other outputs can be more research-oriented, identifying situations that were thought to be good recommendations but in fact may not be, according to decisions and reasons explicitly provided by physicians while using the system.
The reader may have noted that we are not emphasizing from the start that the requirements of the data sets generated by our approach include being of considerable size (the "V" for “volume”). The reason for this is that we are convinced that the data generated will eventually grow. However, there is an increasing need to prioritize the "V" for “value.” We think this value is closely linked to ensuring the "V" for “veracity” in big data approaches in healthcare, beyond the rest of the Vs (velocity, variety), that are certainly depending on technological capabilities and solutions. This means that we need to ensure mechanisms to guarantee the quality and completeness of the data collected[31][32] in normalized repositories if we want to have success in applying these techniques and obtaining valuable healthcare results.
Conclusion
Decision support systems might be able to facilitate the autonomy of citizens when choosing their health options and the ability of professionals to make the most appropriate decision at the right moment. It may also help health policy makers and managers to prioritize the most needed actions in an environment with increasing health needs and resource constraints. But this will be very difficult without the development and maintenance of repositories of dissociated and normalized relevant clinical data from the daily clinical practice, the contributions of the patients themselves, and the fusion with open-access data of the social environment. Furthermore, this should be quickly accompanied by a proper regulation[33] (by the qualified bodies in Europe and the FDA in the U.S.) that make clearer for entrepreneurs the requirements for the development, testing, and validation of these new models.
References
- ↑ Institute of Medicine (1990). Field, M.J.; Lohr, K.N.. ed. Clinical Practice Guidelines: Directions for a New Program. National Academies Press. doi:10.17226/1626.
- ↑ 2.0 2.1 Moskowitz, A.; McSparron, J.; Stone, D.J.; Celi, L.A. (2015). "Preparing a New Generation of Clinicians for the Era of Big Data". Harvard Medical Student Review 2 (1): 24–27. PMC PMC4327872. PMID 25688383. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4327872.
- ↑ Toussi, M.; Lamy, J.B.; Le Toumelin, P.; Venot, A. (2009). "Using data mining techniques to explore physicians' therapeutic decisions when clinical guidelines do not provide recommendations: Methods and example for type 2 diabetes". BMC Medical Informatics and Decision Making 9: 28. doi:10.1186/1472-6947-9-28. PMC PMC2700100. PMID 19515252. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2700100.
- ↑ Gabbay, J.; le May, A. (2004). "Evidence based guidelines or collectively constructed "mindlines?": Ethnographic study of knowledge management in primary care". BMJ 329 (7473): 1013. doi:10.1136/bmj.329.7473.1013. PMC PMC524553. PMID 15514347. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC524553.
- ↑ Handler, T.; Hieb, B. (13 June 2007). "The Updated Gartner CPR Generation Criteria" (PDF). Gartner Teleconference. Gartner. https://hiriresearch.files.wordpress.com/2011/04/cpr-generational-model.pdf.
- ↑ Greenes, R.A. (2015). "Evolution and Revolution in Knowledge-Driven Health IT: A 50-Year Perspective and a Look Ahead". In Riaño, D.; Lenz, R.; Miksch, S. et al.. Knowledge Representation for Health Care. Lecture Notes in Computer Science. 9485. Springer. pp. 3–20. doi:10.1007/978-3-319-26585-8_1. ISBN 9783319265858.
- ↑ Wang, D.; Peleg, M.; Tu, S.W. et al. (2004). "Design and implementation of the GLIF3 guideline execution engine". Journal of Biomedical Informatics 37 (5): 305–18. doi:10.1016/j.jbi.2004.06.002. PMID 15488745.
- ↑ Peleg, M. (2013). "Computer-interpretable clinical guidelines: A methodological review". Journal of Biomedical Informatics 46 (4): 744–63. doi:10.1016/j.jbi.2013.06.009. PMID 23806274.
- ↑ González-Ferrer, A.; Peleg, M. (2015). "Understanding requirements of clinical data standards for developing interoperable knowledge-based DSS: A case study". Computer Standards & Interfaces 42: 125–36. doi:10.1016/j.csi.2015.06.002.
- ↑ 10.0 10.1 Parimbelli, E.; Sacchi, L.; Bellazzi, R. (2016). "Decision Support through Data Integration: Strategies to Meet the Big Data Challenge". European Journal for Biomedical Informatics 12 (1): en10–en14. https://www.ejbi.org/archive/ejbi-volume-12-issue-1-year-2016.html.
- ↑ Mandel, J.C.; Kreda, D.A.; Mandl, K.D. et al. (2016). "SMART on FHIR: A standards-based, interoperable apps platform for electronic health records". JAMIA 23 (5): 899-908. doi:10.1093/jamia/ocv189. PMC PMC4997036. PMID 26911829. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4997036.
- ↑ Peleg, M.; Shahar, Y.; Quaglini, S. (2013). "Making healthcare more accessible, better, faster, and cheaper: The MobiGuide Project". European Journal of ePractice (20): 5–20. https://joinup.ec.europa.eu/sites/default/files/document/2014-06/ePractice%20Journal-Vol.20-November%202013.pdf.
- ↑ Lenz, R.; Peleg, M.; Reichert, M. (2012). "Healthcare Process Support: Achievements, Challenges, Current Research". International Journal of Knowledge-Based Organizations 2 (4): i–xvi. https://www.igi-global.com/journal/international-journal-knowledge-based-organizations/1177.
- ↑ van der Aalst, W.; Adriansyah, A.; Alves de Medeiros, A.K. et al. (2012). "Process Mining Manifesto". In Daniel, F.; Barkaoui, K.; Dustdar, S.. Business Process Management Workshops 2011. Lecture Notes in Business Information Processing. 99. Springer. doi:10.1007/978-3-642-28108-2_19. ISBN 9783642281082.
- ↑ Mans, R.S.; van der Aalst, W.; Vanwersch, R.J.B. et al. (2013). "Process Mining in Healthcare: Data Challenges When Answering Frequently Posed Questions". In Lenz, R.; Miksch, S.; Peleg, M. et al.. Process Support and Knowledge Representation in Health Care. Lecture Notes in Computer Science. 7738. Springer. doi:10.1007/978-3-642-36438-9_10. ISBN 9783642364389.
- ↑ Bellazzi, R.; Zupan, B. (2008). "Predictive data mining in clinical medicine: Current issues and guidelines". International Journal of Medical Informatics 77 (2): 81–97. doi:10.1016/j.ijmedinf.2006.11.006. PMID 17188928.
- ↑ Bellazzi, R.; Ferrazzi, F.; Sacchi, L. (2011). "Predictive data mining in clinical medicine: A focus on selected methods and applications". Wiley Interdisciplinary Reviews: Data Mining and Knowledge Discovery 1 (5): 416–30. doi:10.1002/widm.23.
- ↑ Dawson-Saunders, B.; Feltovich, P.J.; Coulson, R.L.; Steward, D.E. (1990). "A survey of medical school teachers to identify basic biomedical concepts medical students should understand". Academic Medicine 65 (7): 448–54. PMID 2242199.
- ↑ González-Ferrer, A.; Valcárcel, M.; Cháfer, J. et al. (2016). "Diagnóstico y tratamiento de hiponatremia usando modelos computacionales de guías de práctica clínica". Actas del XIX Congreso Nacional de Informática para la Salud, INFORSALUD 2016: 193–198.
- ↑ González-Ferrer, A.; Valcárcel, M.; Cuesta, M. et al. (2017). "Development of a computer-interpretable clinical guideline model for decision support in the differential diagnosis of hyponatremia". International Journal of Medical Informatics 103: 55–64. doi:10.1016/j.ijmedinf.2017.04.014. PMID 28551002.
- ↑ Fox, J.; Johns, N.; Rahmanzadeh, A. et al. (1998). "Disseminating medical knowledge: The PROforma approach". Artificial Intelligence in Medicine 14 (1–2): 157–81. PMID 9779888.
- ↑ Fox, J.; Gutenstein, M.; Khan, O. et al. (2015). "OpenClinical.net: A platform for creating and sharing knowledge and promoting best practice in healthcare". Computers in Industry 66: 63–72. doi:10.1016/j.compind.2014.10.001.
- ↑ Fenske, W.; Maier, S.K.; Blechschmidt, A. et al. (2010). "Utility and limitations of the traditional diagnostic approach to hyponatremia: A diagnostic study". American Journal of Medicine 123 (7): 652–7. doi:10.1016/j.amjmed.2010.01.013. PMID 20609688.
- ↑ González-Ferrer, A.; Valcárcel, M.; Cuesta, M. et al.. "Comorbidities in the Syndrome of Inappropriate Antidiuretic Hormone Secretion: A Hierarchical Clustering Analysis on Discharge Data". To be published.
- ↑ Clarke, K.R.; Somerfield, P.J.; Gorley, R.N. et al. (2008). "Testing of null hypotheses in exploratory community analyses: Similarity profiles and biota-environment linkage". Journal of Experimental Marine Biology and Ecology 366 (1–2): 56–69. doi:10.1016/j.jembe.2008.07.009.
- ↑ Owen, S; Anil, R.; Dunning, T. et al. (2011). Mahout in Action. Manning Publications. pp. 416. ISBN 9781935182689.
- ↑ "Smart Hospitals: Security and Resilience for Smart Health Service and Infrastructures". European Union Agency for Network and Information Security. November 2016. doi:10.2824/28801. https://www.enisa.europa.eu/publications/cyber-security-and-resilience-for-smart-hospitals.
- ↑ Seara, G.; Payá, A.; Mayol, J. (2016). "Value-based healthcare delivery in the digital era". European Psychiatry 33 (Supplement): S33. doi:10.1016/j.eurpsy.2016.01.862.
- ↑ Gutiérrez, A.R.; Cuenca, G.M.; Acebedo, I.A. et al. (June 2013). [http://gesdoc.isciii.es/gesdoccontroller?action=download&id=29/11/2013-45c9ee530c "Manual práctico de interoperabilidad semántica para entornos sanitarios basada en arquetipos"] (PDF). Unidad de Investigación en Telemedicina y e-Salud. pp. 152. http://gesdoc.isciii.es/gesdoccontroller?action=download&id=29/11/2013-45c9ee530c.
- ↑ "Regulation (EU) 2016/679 of the European Parliament and of the Council of 27 April 2016 on the protection of natural persons with regard to the processing of personal data and on the free movement of such data, and repealing Directive 95/46/EC (General Data Protection Regulation)". EUR-Lex. European Union. 2016. http://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:32016R0679.
- ↑ Costa-Pereira, A.; Chen, R.; Almeida, F.C. et al. (2009). "Chapter 4: Data Quality and Integration Issues in Electronic Health Records". In Hristidis, V.. Information Discovery on Electronic Health Records. Taylor & Francis Group. pp. 55–95. ISBN 9781420090413.
- ↑ Weiskopf, N.G.; Weng, C. (2013). "Methods and dimensions of electronic health record data quality assessment: Enabling reuse for clinical research". JAMIA 20 (1): 144–51. doi:10.1136/amiajnl-2011-000681. PMC PMC3555312. PMID 22733976. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3555312.
- ↑ Brown, S.H.; Miller, R.A. (2014). "Chapter 26: Legal and Regulatory Issues Related to the Use of Clinical Software in Health Care Delivery". In Greenes, R.A.. Clinical Decision Support. Academic Press. pp. 711–740. doi:10.1016/B978-0-12-398476-0.00026-9. ISBN 9780123984760.
Notes
This presentation is faithful to the original, with only a few minor changes to grammar, spelling, and presentation, including the addition of PMCID and DOI when they were missing from the original reference. The inline citation for citation 24 was misnumbered in the original text; it's corrected here.