Journal:Information technology support for clinical genetic testing within an academic medical center
| Full article title | Information technology support for clinical genetic testing within an academic medical center |
|---|---|
| Journal | Journal of Personalized Medicine |
| Author(s) | Aronson, Samuel; Mahanta, Lisa; Ros, Lei L.; Clark, Eugene; Babb, Lawrence; Oates, Michael; Rehm, Heidi; Lebo, Matthew |
| Author affiliation(s) | Partners HealthCare, Brigham & Women’s Hospital, Harvard Medical School, Broad Institute of Massachusetts Institute of Technology and Harvard |
| Primary contact | Email: saronson at partners dot org |
| Editors | Liggett, Stephen B. |
| Year published | 2016 |
| Volume and issue | 6 (1) |
| Page(s) | 4 |
| DOI | 10.3390/jpm6010004 |
| ISSN | 2075-4426 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | http://www.mdpi.com/2075-4426/6/1/4 |
| Download | http://www.mdpi.com/2075-4426/6/1/4/pdf (PDF) |
Abstract
Academic medical centers require many interconnected systems to fully support genetic testing processes. We provide an overview of the end-to-end support that has been established surrounding a genetic testing laboratory within our environment, including both laboratory and clinician-facing infrastructure. We explain key functions that we have found useful in the supporting systems. We also consider ways that this infrastructure could be enhanced to enable deeper assessment of genetic test results in both the laboratory and clinic.
Keywords: genetic testing process, information technology support, LIMS, LIS, GeneInsight, GIGPAD
Introduction
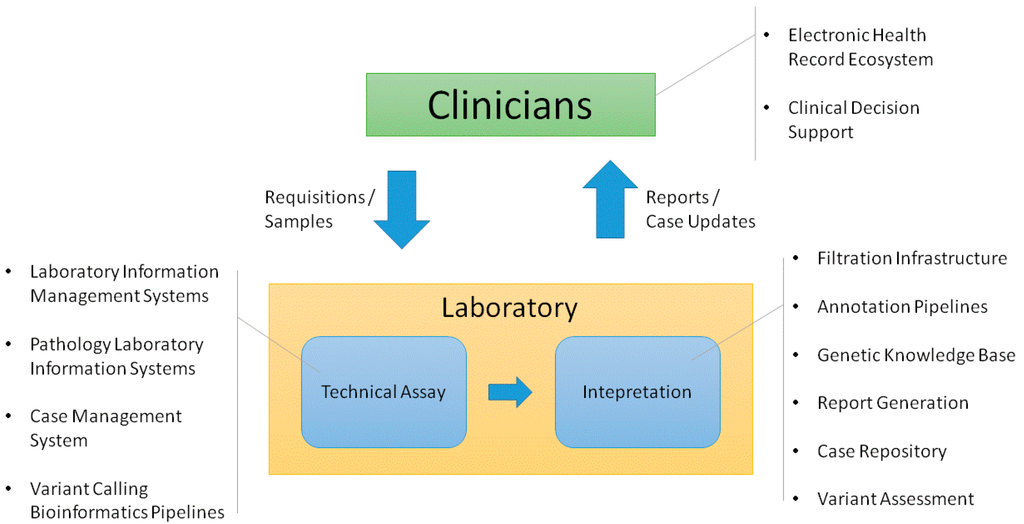
The clinical genetic testing process within an academic medical center involves multiple different groups. Clinicians identify the need for tests and order them. Then, laboratory technicians receive these orders with associated samples and perform the requested assays. The results of these assays are sent to laboratory personnel responsible for interpreting the results and ultimately producing a report that is signed out by a board certified professional. This report is then returned to the clinician who is responsible for assessing how the results should affect the patient’s care. The clinician shares the assessment with the patient and also stores and manages the results over time.[1]
Each of the groups involved in this process requires information technology (IT) support to perform its function in a high quality, efficient manner. This often involves establishing deep support for specific processes. Multiple systems may be needed to meet a group’s needs. In this context, it is important to determine how systems should be integrated across the workflow. The quality of the overall clinical process depends on data transferring in a robust, consistent manner. Figure 1 shows the infrastructure that we have found necessary to support these process flows in the germline genetic testing environment. There are multiple ways this functionality can be implemented. While a complete review of different methods is beyond the scope of this article, we will describe the infrastructure that we have established to support the clinical germline genetic testing processes implemented by our Laboratory for Molecular Medicine (LMM).
|
Clinician-facing infrastructure
The electronic health record (EHR) is the basis for most clinical IT support. However, the EHR often has to be extended to provide in-depth support for a particular area or function.[2] As a result, it is useful to think in terms of an EHR ecosystem that includes other systems linked to the EHR. We have found that the clinical genetic IT support needed most by clinicians includes helping them receive and manage genetic test results as well as enabling them to stay up to date on changing genetic knowledge.
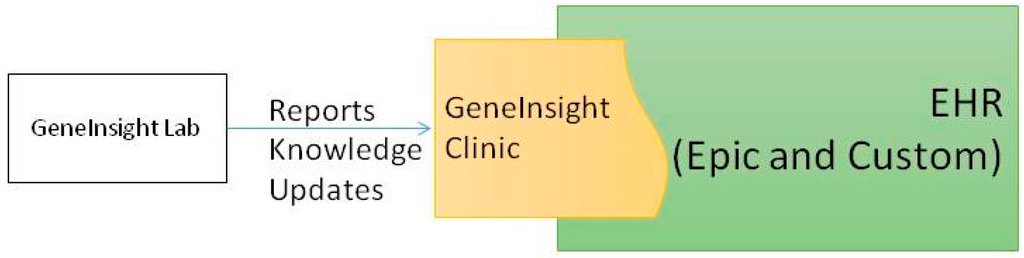
Our institution is in the process of transitioning from a custom built EHR to Epic (Epic Systems, Verona, WI, USA). We have integrated an external application we created, GeneInsight Clinic[3], with both of these EHRs to provide genetics support for our clinicians (Figure 2). GeneInsight Clinic provides clinicians access to easily view genetic test results that have been executed on a patient by our LMM and the potentially meaningful variants that were identified by these tests by incorporating structured data delivered by the molecular genetics laboratory. Users can then drill down into the interpretation of those variants that were assessed relative to the indication for testing. It also provides clinicians with alerts when one of these interpretations is updated by the reporting lab in a potentially clinically significant way.[4] We also recognize the need for additional forms of clinical decision support (CDS)[5] including rules being developed by the National Academy of Medicine’s Display and Integrating Genetic Information Through the EHR Action Collaborative (DIGITizE AC) organized under the genomics roundtable. We have not yet leveraged Substitutable Medical Applications and Reusable Technology (SMART)[5] apps to extend our clinical genetic displays, but we believe this platform has great potential and are investigating doing so.
|
Currently, order and phenotypic information is transferred to the laboratory via paper requisition forms. An electronic order entry system could provide far deeper support, especially for exome and genome based assays. However, there is a challenge in that our laboratory receives orders from around the world. An electronic order entry system would need to be broadly accessible to have the maximum impact and ideally integrated with as many EHRs and laboratory information systems (LIS) as possible. In advance of such functionality becoming available, we have begun to investigate alternate approaches focused on providing the laboratory with access to EHR data when tests are ordered for patients within our environment.
Infrastructure supporting the technical assay
The technical assay workflow
The technical workflow for each assay functions to identify variants present in the patient’s DNA associated to that test. From an IT perspective, it is important to support both the development of these tests and their execution for each patient. Developing tests often involves designing primary assays that broadly assess DNA either within regions of interest or across the whole exome or genome. Often, many technical platforms are required for a clinical test to be complete due to a few factors: (1) technical limitations of the assay requires an alternate platform, (2) confirmation of variants are required using an orthogonal technology, and (3) additional assays are required for completeness (i.e., “fill-in” of the specific regions defined within the test).
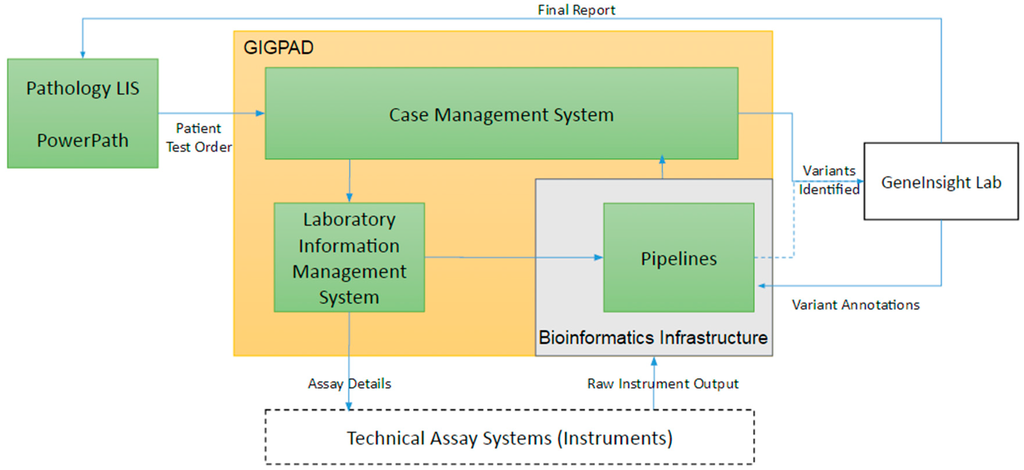
For all of these types of assays, bioinformatic pipelines are needed to process raw instrument output and convert it to variant calls and completeness and coverage of the tested region. In addition to data management, support is also needed to manage the large inventory of oligonucleotides (oligos) required to perform the DNA enrichment and isolation that underlie these assays. Once assays transition into production, laboratories require workflow support to track patients’ test details, manage experiment batch details, organize bioinformatics pipeline runs, record laboratory quality controls, store technical reviews, and consolidate information into case level summaries. Our support consists of three primary systems that are interconnected: PowerPath Pathology laboratory information system (LIS), our Gateway for Integrated Genomics Proteomics Applications and Data (GIGPAD) system and our bioinformatic pipeline infrastructure (Figure 3). GIGPAD in turn serves as enterprise infrastructure that incorporates our Laboratory Information Management Systems (LIMS). We designed GIGPAD so that each lab can make an independent decision whether to custom build a laboratory information management system (LIMS) or buy a commercial solution. Either choice can be integrated into the GIGPAD “superstructure” to support coordinated process flows and piggyback on GIGPAD’s interfaces to other systems. (We use the term LIS to refer to enterprise pathology systems and the term LIMS to refer to systems focused on managing specific wet bench workflows surrounding instruments.) GIGPAD also contains a Case Management System (CMS).
|
References
- ↑ Aronson, S.J.; Rehm, H.L. (2015). "Building the foundation for genomics in precision medicine". Nature 526 (7573): 336–42. doi:10.1038/nature15816. PMID 26469044.
- ↑ Starren, J.; Williams, M.S.; Bottinger, E.P. (2013). "Crossing the omic chasm: A time for omic ancillary systems". JAMA 309 (12): 1237–8. doi:10.1001/jama.2013.1579. PMC PMC3857698. PMID 23494000. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3857698.
- ↑ Aronson, S.J.; Clark, E.H.; Babb, L.J. et al. (2011). "The GeneInsight Suite: a platform to support laboratory and provider use of DNA-based genetic testing". Human Mutation 32 (5): 532–6. doi:10.1002/humu.21470. PMC PMC3082613. PMID 21432942. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3082613.
- ↑ Aronson, S.J.; Clark, E.H.; Varugheese, M. et al. (2012). "Communicating new knowledge on previously reported genetic variants". Genetics in Medicine 14: 713–719. doi:10.1038/gim.2012.19. PMC PMC3841913. PMID 22481129. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3841913.
- ↑ 5.0 5.1 Hoffman, M.A.; Williams, M.S. (2011). "Electronic medical records and personalized medicine". Human Genetics 130 (1): 33–9. doi:10.1007/s00439-011-0992-y. PMID 21519832.
Cite error: Invalid
<ref>tag; name "HoffmanElectro11" defined multiple times with different content
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In several cases the PubMed ID was missing and was added to make the reference more useful.