Difference between revisions of "Journal:PathEdEx – Uncovering high-explanatory visual diagnostics heuristics using digital pathology and multiscale gaze data"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) (Saving and adding more.) |
||
| Line 15: | Line 15: | ||
|doi = [http://doi.org/10.4103/jpi.jpi_29_17 10.4103/jpi.jpi_29_17] | |doi = [http://doi.org/10.4103/jpi.jpi_29_17 10.4103/jpi.jpi_29_17] | ||
|issn = 2153-3539 | |issn = 2153-3539 | ||
|license = [http://www.jpathinformatics.org/article.asp?issn=2153-3539;year=2017;volume=8;issue=1;spage=29;epage=29;aulast=Shin http://www.jpathinformatics.org] | |license = [http://creativecommons.org/licenses/by-nc-sa/3.0/ Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported] | ||
|website = [http://www.jpathinformatics.org/article.asp?issn=2153-3539;year=2017;volume=8;issue=1;spage=29;epage=29;aulast=Shin http://www.jpathinformatics.org] | |||
|download = [http://www.jpathinformatics.org/temp/JPatholInform8129-6283361_172713.pdf http://www.jpathinformatics.org/temp/JPatholInform8129-6283361_172713.pdf] (PDF) | |download = [http://www.jpathinformatics.org/temp/JPatholInform8129-6283361_172713.pdf http://www.jpathinformatics.org/temp/JPatholInform8129-6283361_172713.pdf] (PDF) | ||
}} | }} | ||
| Line 44: | Line 45: | ||
In this paper, we present PathEdEx, a web-based, WSI- and gaze tracking-enabled informatics framework for training and mining of visual and non-visual heuristics of pathology diagnosis. The PathEdEx system enables the development of interactive online atlases that can be used not only for educational purposes but also have the potential of capturing best diagnostic strategies and representing them in a highly explanatory format for pathology practice. We demonstrate the capabilities of PathEdEx for mining visual and non-visual diagnostic heuristics by 1. performing quantitative studies on the time dynamics of zooming and panning operations utilized by experts and novices to come to a correct diagnosis, 2. scanning association rule (AR) mining<ref name="AgrawalMining93">{{cite journal |title=Mining association rules between sets of items in large databases |journal=ACM SIGMOD Record |author=Agrawal, R.; Imieliński, T.; Swami, A. |volume=22 |issue=2 |pages=207–216 |year=1993 |doi=10.1145/170036.170072}}</ref> studies to determine sets of diagnostic factors that consistently result in a correct diagnosis, and 3. quantifying differences in diagnostic strategies across different levels of pathology expertise using Markov chain (MC) modeling<ref name="MarkovExtension07">{{cite book |chapter=Appendix B: Extension of the Limit Theorems of Probability Theory to a Sum of Variables Connected in a Chain |title=Dynamic Probabilistic Systems, Volume I: Markov Models |author=Markov, A. |editor=Howard, R.A. |publisher=Dover Publications |pages=551–576 |year=2007 |isbn=9780486458700}}</ref><ref name="MarkovExtension71">{{cite book |chapter=Extension of the Limit Theorems of Probability Theory to a Sum of Variables Connected in a Chain |title=Dynamic Probabilistic Systems, Volume I: Markov Models |author=Markov, A. |editor=Howard, R.A. |publisher=John Wiley & Sons, Inc |pages=552–577 |year=1971 |isbn=9780471416654}}</ref> and MC Monte Carlo (MCMC) simulations.<ref name="MetropolisEquation53">{{cite journal |title=Equation of State Calculations by Fast Computing Machines |journal=The Journal of Chemical Physics |author=Metropolis, N.; Rosenbluth, A.W.; Rosenbluth, M.N. et al. |volume=21 |pages=1087–1092 |year=1953 |doi=10.1063/1.1699114}}</ref><ref name="RobertAShort11">{{cite journal |title=A Short History of Markov Chain Monte Carlo: Subjective Recollections from Incomplete Data |journal=Statistical Science |author=Robert, C.; Casella, G. |volume=26 |issue=1 |pages=102–115 |year=2011 |doi=10.1214/10-STS351}}</ref> These studies allowed us to understand effective and efficient practices of human diagnostic heuristics that can be passed over to the next generation of pathologists and streamline implementation of precision diagnostics in precision medicine settings. The studies are performed using the first PathEdEx interactive training atlas of hematopathological cancers. | In this paper, we present PathEdEx, a web-based, WSI- and gaze tracking-enabled informatics framework for training and mining of visual and non-visual heuristics of pathology diagnosis. The PathEdEx system enables the development of interactive online atlases that can be used not only for educational purposes but also have the potential of capturing best diagnostic strategies and representing them in a highly explanatory format for pathology practice. We demonstrate the capabilities of PathEdEx for mining visual and non-visual diagnostic heuristics by 1. performing quantitative studies on the time dynamics of zooming and panning operations utilized by experts and novices to come to a correct diagnosis, 2. scanning association rule (AR) mining<ref name="AgrawalMining93">{{cite journal |title=Mining association rules between sets of items in large databases |journal=ACM SIGMOD Record |author=Agrawal, R.; Imieliński, T.; Swami, A. |volume=22 |issue=2 |pages=207–216 |year=1993 |doi=10.1145/170036.170072}}</ref> studies to determine sets of diagnostic factors that consistently result in a correct diagnosis, and 3. quantifying differences in diagnostic strategies across different levels of pathology expertise using Markov chain (MC) modeling<ref name="MarkovExtension07">{{cite book |chapter=Appendix B: Extension of the Limit Theorems of Probability Theory to a Sum of Variables Connected in a Chain |title=Dynamic Probabilistic Systems, Volume I: Markov Models |author=Markov, A. |editor=Howard, R.A. |publisher=Dover Publications |pages=551–576 |year=2007 |isbn=9780486458700}}</ref><ref name="MarkovExtension71">{{cite book |chapter=Extension of the Limit Theorems of Probability Theory to a Sum of Variables Connected in a Chain |title=Dynamic Probabilistic Systems, Volume I: Markov Models |author=Markov, A. |editor=Howard, R.A. |publisher=John Wiley & Sons, Inc |pages=552–577 |year=1971 |isbn=9780471416654}}</ref> and MC Monte Carlo (MCMC) simulations.<ref name="MetropolisEquation53">{{cite journal |title=Equation of State Calculations by Fast Computing Machines |journal=The Journal of Chemical Physics |author=Metropolis, N.; Rosenbluth, A.W.; Rosenbluth, M.N. et al. |volume=21 |pages=1087–1092 |year=1953 |doi=10.1063/1.1699114}}</ref><ref name="RobertAShort11">{{cite journal |title=A Short History of Markov Chain Monte Carlo: Subjective Recollections from Incomplete Data |journal=Statistical Science |author=Robert, C.; Casella, G. |volume=26 |issue=1 |pages=102–115 |year=2011 |doi=10.1214/10-STS351}}</ref> These studies allowed us to understand effective and efficient practices of human diagnostic heuristics that can be passed over to the next generation of pathologists and streamline implementation of precision diagnostics in precision medicine settings. The studies are performed using the first PathEdEx interactive training atlas of hematopathological cancers. | ||
==Methods== | |||
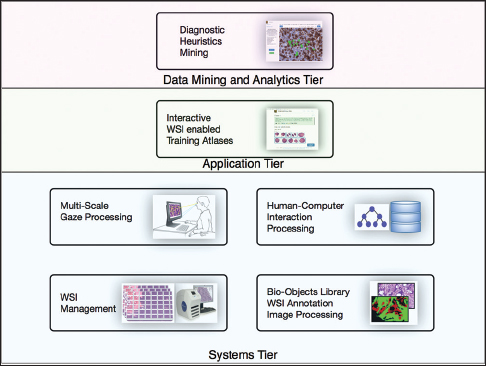
Each PathEdEx training atlas is a web-based system and runs inside a browser window as a thin client in an in-house developed multi-tiered informatics ecosystem, depicted in Figure 1. The systems tier represents a foundation of PathEdEx, containing essential software modules for processing of imaging and non-imaging data. | |||
[[File:Fig1 Shin JofPathInformatics2017 8.jpg|486px]] | |||
{{clear}} | |||
{| | |||
| STYLE="vertical-align:top;"| | |||
{| border="0" cellpadding="5" cellspacing="0" width="486px" | |||
|- | |||
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 1:''' PathEdEx is a multitiered informatics ecosystem that enables education and research in digital pathology and pathology informatics.</blockquote> | |||
|- | |||
|} | |||
|} | |||
===Whole slide imaging management module=== | |||
This module is responsible for processing of WSIs. The client side of this module is implemented in JavaScript using Bootstrap<ref name="BootstrapHome">{{cite web |url=http://getbootstrap.com/ |title=Bootstrap |author=Otto, M.; Thornton, J. |accessdate=23 June 2017}}</ref> and jQuery<ref name="jQueryHome">{{cite web |url=http://jquery.com/ |title=jQuery |publisher=jQuery Foundation |accessdate=23 June 2017}}</ref> libraries, and the WSI online viewer is based on the OpenSeaDragon JavaScript library<ref name="OSHome">{{cite web |url=https://openseadragon.github.io/ |title=Open Seadragon |author=OpenSeadragon contributors |publisher=CodePlex Foundation |accessdate=23 June 2017}}</ref> that enables the interactive viewing of large WSIs over the internet. At the server side, WSIs are served by IIPImage server, which is an open-source tiled image server.<ref name="PillayIIPImage">{{cite web |url=http://iipimage.sourceforge.net/ |title=IIPImage |author=Pillay, R. |publisher=Slashdot Media |accessdate=23 June 2017}}</ref> The WSI images of the patient cases are obtained by scanning the slides with the Aperio ScanScope CS (Leica Biosystems)<ref name="LeicaHome">{{cite web |url=http://www.leicabiosystems.com/ |title=Leica Biosystems |publisher=Leica Biosystems Nussloch GmbH |accessdate=23 June 2017}}</ref> slide scanner at × 40, and they are converted to tiled pyramid TIFF images using the open-source VIPS library.<ref name="CupittVIPS">{{cite web |url=http://www.vips.ecs.soton.ac.uk/index.php?title=VIPS |title=VIPS |author=Cupitt, J.; Martinez, K.; Padfield, J. |accessdate=23 June 2017}}</ref> Each TIFF image contains 256 × 256 pixel tiles of the slide at seven different zoom levels. This enables the efficient communication of the whole slide data since only the visible part of the WSI is served to the client at any time. The WSI online viewer allows users to zoom and pan the image to provide a virtual microscope experience. | |||
==References== | ==References== | ||
Revision as of 23:20, 26 July 2017
| Full article title | PathEdEx – Uncovering high-explanatory visual diagnostics heuristics using digital pathology and multiscale gaze data |
|---|---|
| Journal | Journal of Pathology Informatics |
| Author(s) | Shin, Dmitriy; Kovalenko, Mikhail; Ersoy, Ilker; Li, Yu; Doll, Donald; Shyu, Chi-Ren; Hammer, Richard |
| Author affiliation(s) | University of Missouri – Columbia |
| Primary contact | Email: Available w/ login |
| Year published | 2017 |
| Volume and issue | 8 |
| Page(s) | 29 |
| DOI | 10.4103/jpi.jpi_29_17 |
| ISSN | 2153-3539 |
| Distribution license | Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported |
| Website | http://www.jpathinformatics.org |
| Download | http://www.jpathinformatics.org/temp/JPatholInform8129-6283361_172713.pdf (PDF) |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
Background: Visual heuristics of pathology diagnosis is a largely unexplored area where reported studies only provided a qualitative insight into the subject. Uncovering and quantifying pathology visual and non-visual diagnostic patterns have great potential to improve clinical outcomes and avoid diagnostic pitfalls.
Methods: Here, we present PathEdEx, an informatics computational framework that incorporates whole-slide digital pathology imaging with multiscale gaze-tracking technology to create web-based interactive pathology educational atlases and to datamine visual and non-visual diagnostic heuristics.
Results: We demonstrate the capabilities of PathEdEx for mining visual and non-visual diagnostic heuristics using the first PathEdEx volume of a hematopathology atlas. We conducted a quantitative study on the time dynamics of zooming and panning operations utilized by experts and novices to come to the correct diagnosis. We then performed association rule mining to determine sets of diagnostic factors that consistently result in a correct diagnosis, and studied differences in diagnostic strategies across different levels of pathology expertise using Markov chain (MC) modeling and MC Monte Carlo simulations. To perform these studies, we translated raw gaze points to high-explanatory semantic labels that represent pathology diagnostic clues. Therefore, the outcome of these studies is readily transformed into narrative descriptors for direct use in pathology education and practice.
Conclusion: The PathEdEx framework can be used to capture best practices of pathology visual and non-visual diagnostic heuristics that can be passed over to the next generation of pathologists and have potential to streamline implementation of precision diagnostics in precision medicine settings.
Keywords: Digital pathology, eye tracking, gaze tracking, pathology diagnosis, visual heuristics, visual knowledge, whole slide images
Introduction
Pathology diagnosis is a highly complex process where multiple clinical and diagnostic factors have to be taken into account in an iterative fashion to produce a plausible conclusion that most accurately explains these factors from a biological standpoint.[1] Information from patient clinical history; morphological findings from microscopic evaluation of biopsies, aspirates, and smears; as well as data from flow cytometry, immunohistochemistry (IHC), and "omics" modalities such as comparative hybridization arrays and next-generation sequencing are used in the diagnostic process, which currently can be described more like a subjective exercise than a well-defined protocol. As such, it can frequently lead to diagnostic pitfalls which may harmfully impact patient case with a wrong diagnosis. The diagnostic pitfalls are most often encountered in the case of complex diseases such as cancer[2], where multiple fine-grained clinical phenotypes may require a better understanding of genomic variations across patient populations and therefore require better protocols for pathology diagnosis. It is especially important in light of realizing precision medicine ideas.[3] A better understanding of pathology diagnosis, especially of heuristics of visual reasoning over microscopic slides, is crucial to develop means for new genomic-enabled precision diagnostics methods.
Over the last decade, digital pathology (DP) and whole slide imaging (WSI) have become a mature technology that allows reproduction of the histopathologic glass slide in its entirety.[4] For the first time in the history of microscopy, diagnostic-quality digital images can be stored electronically and analyzed using computer algorithms to assist primary diagnosis and streamline research in biomedical imaging informatics.[5] This was not possible before the DP era, when cropped image areas from pathology slides could be used only to seek second opinions or share the diagnostic details with clinicians. At the same time, gaze-capturing devices have undergone transformation from bulky systems to portable, mobile trackers that can be installed on laptops and used to seamlessly collect a user's gaze.
There have been a number of research studies where WSI was used to analyze visual patterns of regions of interests (ROIs) annotated without[6][7][8][9] and with gaze-tracking technology.[10] In the former case, ROIs were identified either manually or using a viewport analysis, and in the latter case, gaze fixation points were used for the same purpose. Gaze tracking, along with mouse movement, has also been used to study pathologists' attention while viewing WSI during pathology diagnosis[11] as well as to study visual and cognitive aspects of pathology expertise.[12] The underlying idea in the majority of these studies was to analyze captured gaze data with respect to the ROIs marked in the pathology images, gaze time spent within the ROIs, and total number of fixations within the ROIs as measures of diagnostically relevant viewing behavior. However, it is challenging to use ROIs to articulate underlying biology for understanding the visual heuristics of specific diagnostic decisions. It is not trivial to encode morphological patterns of an ROI using narrative language. As such, ROI might not be an effective means to translate best practices findings from gaze-tracking studies into pathology education and pave a road toward precision diagnostics.
In this paper, we present PathEdEx, a web-based, WSI- and gaze tracking-enabled informatics framework for training and mining of visual and non-visual heuristics of pathology diagnosis. The PathEdEx system enables the development of interactive online atlases that can be used not only for educational purposes but also have the potential of capturing best diagnostic strategies and representing them in a highly explanatory format for pathology practice. We demonstrate the capabilities of PathEdEx for mining visual and non-visual diagnostic heuristics by 1. performing quantitative studies on the time dynamics of zooming and panning operations utilized by experts and novices to come to a correct diagnosis, 2. scanning association rule (AR) mining[13] studies to determine sets of diagnostic factors that consistently result in a correct diagnosis, and 3. quantifying differences in diagnostic strategies across different levels of pathology expertise using Markov chain (MC) modeling[14][15] and MC Monte Carlo (MCMC) simulations.[16][17] These studies allowed us to understand effective and efficient practices of human diagnostic heuristics that can be passed over to the next generation of pathologists and streamline implementation of precision diagnostics in precision medicine settings. The studies are performed using the first PathEdEx interactive training atlas of hematopathological cancers.
Methods
Each PathEdEx training atlas is a web-based system and runs inside a browser window as a thin client in an in-house developed multi-tiered informatics ecosystem, depicted in Figure 1. The systems tier represents a foundation of PathEdEx, containing essential software modules for processing of imaging and non-imaging data.
|
Whole slide imaging management module
This module is responsible for processing of WSIs. The client side of this module is implemented in JavaScript using Bootstrap[18] and jQuery[19] libraries, and the WSI online viewer is based on the OpenSeaDragon JavaScript library[20] that enables the interactive viewing of large WSIs over the internet. At the server side, WSIs are served by IIPImage server, which is an open-source tiled image server.[21] The WSI images of the patient cases are obtained by scanning the slides with the Aperio ScanScope CS (Leica Biosystems)[22] slide scanner at × 40, and they are converted to tiled pyramid TIFF images using the open-source VIPS library.[23] Each TIFF image contains 256 × 256 pixel tiles of the slide at seven different zoom levels. This enables the efficient communication of the whole slide data since only the visible part of the WSI is served to the client at any time. The WSI online viewer allows users to zoom and pan the image to provide a virtual microscope experience.
References
- ↑ Shin, D.; Arthur, G.; Caldwell, C. et al. (2012). "A pathologist-in-the-loop IHC antibody test selection using the entropy-based probabilistic method". Journal of Pathology Informatics 3: 1. doi:10.4103/2153-3539.93393. PMC PMC3307231. PMID 22439121. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3307231.
- ↑ Higgins, R.A.; Blankenship, J.E.; Kinney, M.C. (2008). "Application of immunohistochemistry in the diagnosis of non-Hodgkin and Hodgkin lymphoma". Archives of Pathology & Laboratory Medicine 132 (3): 441-61. doi:10.1043/1543-2165(2008)132[441:AOIITD]2.0.CO;2. PMID 18318586.
- ↑ Tenenbaum, J.D.; Avillach, P.; Benham-Hutchins, M. et al. (2016). "An informatics research agenda to support precision medicine: Seven key areas". JAMIA 23 (4): 791-5. doi:10.1093/jamia/ocv213. PMC PMC4926738. PMID 27107452. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4926738.
- ↑ Pantanowitz, L. (2010). "Digital images and the future of digital pathology". Journal of Pathology Informatics 1: 15. doi:10.4103/2153-3539.68332. PMC PMC2941968. PMID 20922032. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2941968.
- ↑ Chen, Z.; Shin, D.; Chen, S. et al. (2014). "Histological quantitation of brain injury using whole slide imaging: A pilot validation study in mice". PLOS One 9 (3): e92133. doi:10.1371/journal.pone.0092133. PMC PMC3956884. PMID 24637518. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3956884.
- ↑ Reder, N.P.; Glasser, D.; Dintzis, S.M. et al. (2016). "NDER: A novel web application using annotated whole slide images for rapid improvements in human pattern recognition". Journal of Pathology Informatics 7: 31. doi:10.4103/2153-3539.186913. PMC PMC4977980. PMID 27563490. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4977980.
- ↑ Mercan, E.; Aksoy, S.; Shapiro, L.G. et al. (2016). "Localization of Diagnostically Relevant Regions of Interest in Whole Slide Images: A Comparative Study". Journal of Digital Imaging 29 (4): 496-506. doi:10.1007/s10278-016-9873-1. PMC PMC4942394. PMID 26961982. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4942394.
- ↑ Mercan, C.; Mercan, E.; Aksoy, S. et al. (2016). "Multi-instance multi-label learning for whole slide breast histopathology". SPIE Proceedings 9791: 979108. doi:10.1117/12.2216458.
- ↑ Roa-Peña, L.; Gómez, F.; Romero, E. (2010). "An experimental study of pathologist's navigation patterns in virtual microscopy". Diagnostic Pathology 5: 71. doi:10.1186/1746-1596-5-71. PMC PMC3001424. PMID 21087502. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3001424.
- ↑ Brunyé,T.T.; Carney, P.A.; Allison, K.H. et al. (2014). "Eye movements as an index of pathologist visual expertise: A pilot study". PLOS One 9 (8): e103447. doi:10.1371/journal.pone.0103447. PMC PMC4118873. PMID 25084012. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4118873.
- ↑ Raghunath, V.; Braxton, M.O.; Gagnon, S.A. et al. (2012). "Mouse cursor movement and eye tracking data as an indicator of pathologists' attention when viewing digital whole slide images". Journal of Pathology Informatics 3: 43. doi:10.4103/2153-3539.104905. PMC PMC3551530. PMID 23372984. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3551530.
- ↑ Jaarsma, T; Jarodzka, H.; Nap, M. et al. (2015). "Expertise in clinical pathology: combining the visual and cognitive perspective". Journal of Pathology Informatics 20 (4): 1089-106. doi:10.1007/s10459-015-9589-x. PMC PMC4564442. PMID 25677013. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4564442.
- ↑ Agrawal, R.; Imieliński, T.; Swami, A. (1993). "Mining association rules between sets of items in large databases". ACM SIGMOD Record 22 (2): 207–216. doi:10.1145/170036.170072.
- ↑ Markov, A. (2007). "Appendix B: Extension of the Limit Theorems of Probability Theory to a Sum of Variables Connected in a Chain". In Howard, R.A.. Dynamic Probabilistic Systems, Volume I: Markov Models. Dover Publications. pp. 551–576. ISBN 9780486458700.
- ↑ Markov, A. (1971). "Extension of the Limit Theorems of Probability Theory to a Sum of Variables Connected in a Chain". In Howard, R.A.. Dynamic Probabilistic Systems, Volume I: Markov Models. John Wiley & Sons, Inc. pp. 552–577. ISBN 9780471416654.
- ↑ Metropolis, N.; Rosenbluth, A.W.; Rosenbluth, M.N. et al. (1953). "Equation of State Calculations by Fast Computing Machines". The Journal of Chemical Physics 21: 1087–1092. doi:10.1063/1.1699114.
- ↑ Robert, C.; Casella, G. (2011). "A Short History of Markov Chain Monte Carlo: Subjective Recollections from Incomplete Data". Statistical Science 26 (1): 102–115. doi:10.1214/10-STS351.
- ↑ Otto, M.; Thornton, J.. "Bootstrap". http://getbootstrap.com/. Retrieved 23 June 2017.
- ↑ "jQuery". jQuery Foundation. http://jquery.com/. Retrieved 23 June 2017.
- ↑ OpenSeadragon contributors. "Open Seadragon". CodePlex Foundation. https://openseadragon.github.io/. Retrieved 23 June 2017.
- ↑ Pillay, R.. "IIPImage". Slashdot Media. http://iipimage.sourceforge.net/. Retrieved 23 June 2017.
- ↑ "Leica Biosystems". Leica Biosystems Nussloch GmbH. http://www.leicabiosystems.com/. Retrieved 23 June 2017.
- ↑ Cupitt, J.; Martinez, K.; Padfield, J.. "VIPS". http://www.vips.ecs.soton.ac.uk/index.php?title=VIPS. Retrieved 23 June 2017.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation and updates to spelling and grammar. In some cases important information was missing from the references, and that information was added.