Journal:Sample identifiers and metadata to support data management and reuse in multidisciplinary ecosystem sciences
| Full article title | Sample identifiers and metadata to support data management and reuse in multidisciplinary ecosystem sciences |
|---|---|
| Journal | Data Science Journal |
| Author(s) | Damerow, Joan E.; Varadharajan, Charuleka; Boye, Kristin; Brodie, Eoin L.; Burrus, Madison; Chadwick, K. Dana; Crystal-Ornelas, Robert; Elbashandy, Hesham; Alves, Ricardo J.E.; Ely, Kim S.; Goldman, Amy E.; Haberman, Ted; Hendrix, Valerie; Kakalia, Zarine; Kemner, Kenneth M.; Kersting, Annie B.; Merino, Nancy; O'Brien, Fianna; Perzan, Zach; Robles, Emily; Sorensen, Patrick; Stegen, James C.; Walls, Ramona L.; Weisenhorn, Pamela; Zavarin, Mavrik; Agarwal, Deborah |
| Author affiliation(s) | Lawrence Berkeley National Laboratory, SLAC National Accelerator Laboratory, Stanford University, Brookhaven National Laboratory, Pacific Northwest National Laboratory, Metadata Game Changers, Argonne National Laboratory, Lawrence Livermore National Laboratory, University of Arizona |
| Primary contact | Email: JoanDamerow at lbl dot gov |
| Year published | 2021 |
| Volume and issue | 20(1) |
| Article # | 11 |
| DOI | 10.5334/dsj-2021-011 |
| ISSN | 1683-1470 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://datascience.codata.org/articles/10.5334/dsj-2021-011/ |
| Download | https://datascience.codata.org/articles/10.5334/dsj-2021-011/galley/1055/download/ (PDF) |
Abstract
Physical samples are foundational entities for research across the biological, Earth, and environmental sciences. Data generated from sample-based analyses are not only the basis of individual studies, but can also be integrated with other data to answer new and broader-scale questions. Ecosystem studies increasingly rely on multidisciplinary team-based science to study climate and environmental changes. While there are widely adopted conventions within certain domains to describe sample data, these have gaps when applied in a multidisciplinary context.
In this study, we reviewed existing practices for identifying, characterizing, and linking related environmental samples. We then tested practicalities of assigning persistent identifiers to samples, with standardized metadata, in a pilot field test involving eight United States Department of Energy projects. Participants collected a variety of sample types, with analyses conducted across multiple facilities. We address terminology gaps for multidisciplinary research and make recommendations for assigning identifiers and metadata that supports sample tracking, integration, and reuse. Our goal is to provide a practical approach to sample management, geared towards ecosystem scientists who contribute and reuse sample data.
Keywords: International GeoSample Numbers (IGSN), physical samples, soil, water, plant, leaf, microbial communities, related identifiers, persistent identifiers
Introduction
The study of natural ecosystems requires multidisciplinary science teams to understand and model processes from molecular to global scales.[1] Many research activities involve diverse collections of samples and associated field or laboratory measurements.[2][3] For example, studies of organic matter cycling through plants and soil involves analysis of samples to represent soil biogeochemistry, microbial communities, plant structures, leaf gas exchange, and traits of the specific organisms involved.[4][5][6] Each scientific expert, project team, and discipline has a responsibility to ensure that others can interpret, integrate, and reuse their sample data to help solve emerging problems as our global environment continues to change.[7]
Collaboration across disciplines requires a more unified approach to report basic information about key data entities, such as samples. One challenge in promoting a unified way of reporting sample data is that some research communities have already developed community-specific conventions, including those for omics samples[8][9][10], biodiversity records[11], and geoscience samples.[2][12] A larger challenge is that many researchers use no formal reporting conventions, or exclude information needed to interpret and reuse the data.[13] More coordination is needed across these communities to develop a multidisciplinary reporting format for physical samples that is widely adopted, or to ensure that standards are interoperable. Common reporting would support effective discovery, integration, and reuse of sample data that spans scientific domains.
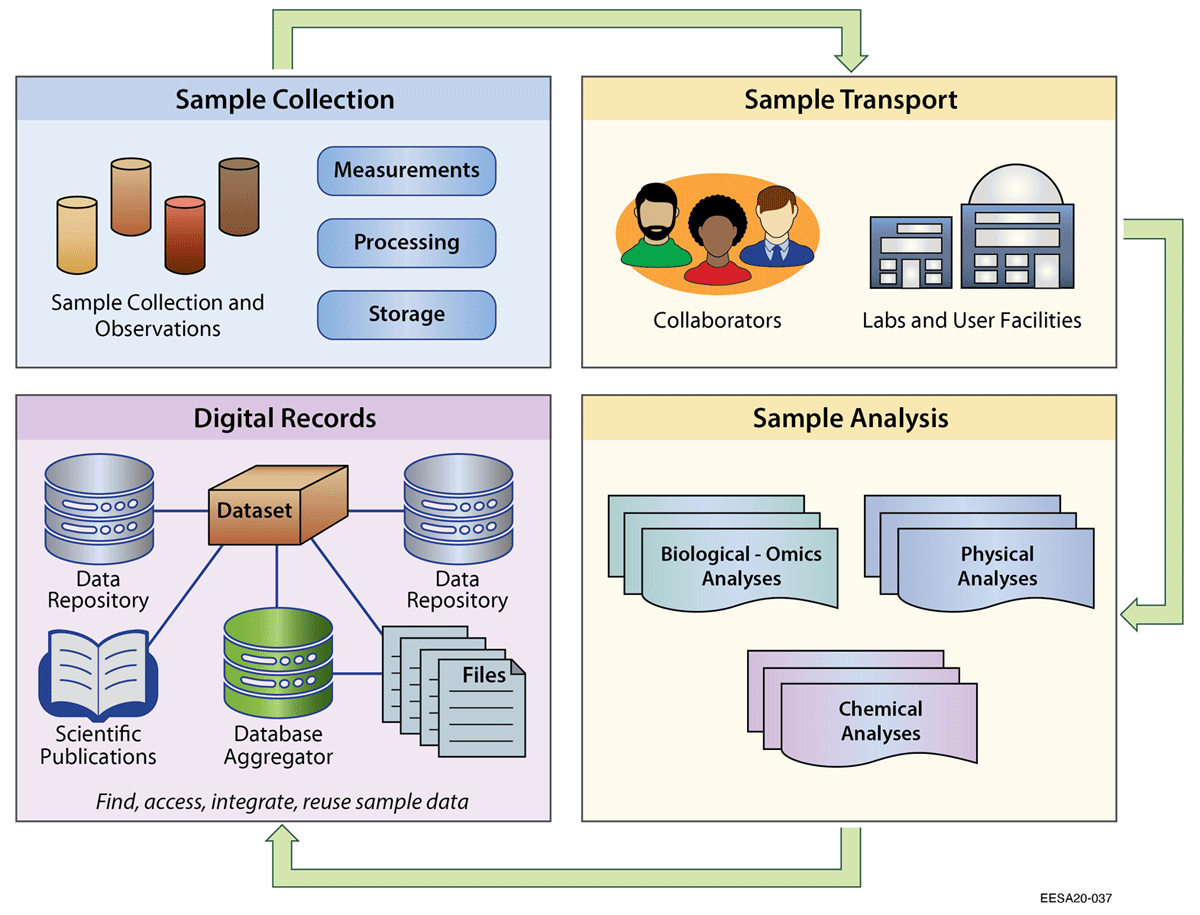
Sample identifiers are also needed to associate and manage important information describing a sample (i.e., metadata), such as the location, date, environmental context, and purpose of sample collection. For multidisciplinary studies, the task of generating and managing unique sample identifiers and associated metadata can be complicated, particularly as important contextual information is added throughout the data lifecycle.[14] Samples are sent to different collaborators, laboratories, and user facilities, and then combined into a variety of digital records and publications (Figure 1).[15] As a result, scientists face challenges with data management, metadata management, tracking, or the ability to integrate and reuse valuable sample data. Without attention, these inefficiencies result in data and metadata loss and inhibit the potential of scientific discovery.
|
Our overall goal was to address sample identification and metadata needs of ecosystem scientists, and was driven by the user community of the U.S. Department of Energy’s (DOE’s) data repository for Earth and environmental sciences, the Environmental Systems Science Data Infrastructure for a Virtual Ecosystem (ESS-DIVE).[16] The DOE’s Environmental Systems Science (ESS) program relies on multidisciplinary, team-based science to study complex processes within terrestrial ecosystems, spanning from the bedrock through the rhizosphere and vegetation to the atmospheric surface layer.[17] This community is well-positioned to help address specific challenges in standardizing and integrating data and metadata about a variety of environmental samples (e.g., soil, water, plant, and associated biological material used for omics analyses), which applies broadly to environmental research.[18][19][20][21][22]
We focus on sample identifiers and metadata that support the FAIR Guiding Principles (findability, accessibility, interoperability, and reusability) from the multidisciplinary domain-science perspective.[23][24][25][26][27] We therefore use a community-focused approach to: a.) evaluate existing options for sample identifiers and metadata descriptions for ecosystem science samples; b.) pilot the process of standardizing sample information to evaluate practical issues from domain-science perspectives; and c.) outline practical recommendations for sample identifier allocation, tracking, and associated metadata.
Methods
Review of existing sample identifiers, metadata conventions, and standards
ESS-DIVE’s work on sample identifiers and metadata began in response to a specific problem with tracking multidisciplinary samples, as they are sent to different labs and user facilities, which DOE ESS scientists brought up during community meetings. As a community-focused data repository, our approach to this issue involved leading or participating in a variety of community discussions on sample identifiers and/or associated metadata. These included:
- presenting identifier options in an ESS community webinar and whitepaper;
- engaging in discussion with each pilot test participant;
- holding several meetings with U.S. DOE user facilities and data systems representatives (Joint Genome Institute, National Microbiome Data Collaborative, Environmental Molecular Sciences Laboratory, and DOE Systems Biology Knowledgebase);
- participating in broader community meetings on identifier and metadata practices for physical samples (Earth Science Information Partners [ESIP] and Research Data Alliance [RDA]]);
- participating in a National Microbiome Data Collaborative (NMDC) Ontology workshop;
- participating in a USGS workshop on sample collection metadata for the National Digital Catalogue; and
- participating in the IGSN 2040 Steering Committee and business planning.
After reviewing the scope and use of available persistent identifier (PID) options (Table 1) and community discussions, we focused additional identifier comparison on International GeoSample Numbers (IGSNs) and Archival Resource Keys (ARKs), which are most commonly used for a variety of sample types (Additional files, Supplemental Table 1). Considerations in the identifier assessment included association with a broader international community focused on sample identification and description, associated metadata to describe samples and their relationships, availability of user-friendly infrastructure to mint identifiers and validate metadata, general ease of use, and other technical identifier characteristics, listed in Additional files, Supplemental Table 1.
| |||||||||||||||||||||||||||||||||
We also reviewed existing metadata standards and templates that are relevant for samples collected by environmental scientists, including general digital object standards[29][30][31], biodiversity records[11][32], omics (e.g. genomics, metagenomics) material[8][10][33], and geoscience samples[12][34] (see Additional files, Supplemental Table 2). We created a translation table comparing 49 metadata elements (see Additional files, Supplemental Table 3) in human-readable format. The translation table depicts linkages where metadata elements were common across standards, as well as differences.
The core IGSN Descriptive Metadata Schema[35] includes basic metadata associated with sample collection, which is generally relevant across sample types. This schema links metadata profiles that differ across six currently-functioning IGSN allocating agents. The System For Earth Sample Registration (SESAR; the first allocating agent) has no access restrictions for obtaining IGSNs and provides user-friendly services for sample management.[36] The SESAR metadata profile and controlled terms are currently focused on geoscience samples, but the IGSN organization seeks to accommodate multiple disciplines and has already expanded into plant and other biological samples for some IGSN allocating agents. Our translation table for sample metadata allowed us to identify metadata elements and terms that could be revised or extended within the SESAR profile for improved representation of other sample types (see Additional files, Supplemental Table 3).
Biology-related standards are well-established, commonly used in the community, and are particularly important for ecosystem science samples. Genomic and metagenomic analyses and data publication require use of standards developed by the Genetic Standards Consortium (GSC)[8], namely Minimum Information about any Sequence (MIxS) and Minimum Information about any Metagenome (MIMS).[10] DarwinCore is a metadata standard for biodiversity records that has been widely adopted across the biocollections community.[11] It is also required for submitting data to the Global Biodiversity Information Facility (GBIF), which allows global search and integration of biodiversity records.[37][38] GBIF provides a valuable service as a data aggregator, and thus has driven standards adoption, enabling a wide range of data reuse applications in published biodiversity studies[37][39], including over 5,000 known citations from studies using biodiversity records.[40]
We researched ontologies that could be used to describe a broad set of environmental sample types, including the Biological Collections Ontology (BCO)[41], Environment Ontology (ENVO)[42], Population and Community Ontology (PCO)[43], and Plant Ontology (PO)[44] to identify additional or alternate terms to generally describe other types of soil, sediment, water, gas, and biology-related samples.[45]
We also engaged with the broader, international community working on sample-related practices. This broader community is led by members of the IGSN organization, with participation across other national agencies (e.g. USGS, CSIRO, Australia Research Data Commons-ARDC) and data organizations (ESIP and RDA). This community participation was important in identifying best practices in identifier and metadata use, and contributing perspectives of ecosystem sciences in the broader community working on sample standardization. Continued participation in the broader informatics and domain science communities is important for improving interoperability and usability of sample-related standards.
Sample identifier and metadata testing in the field
In order to develop a sample metadata reporting format that was informed by our domain science community, we worked with scientists from eight different Environmental Systems Science projects to conduct a pilot test for using sample PIDs and metadata. In particular, we tested the practicality of the IGSN, which appeared to be the best choice amongst relevant PIDs for our purposes. These projects had varying scopes and sample types, and were all funded by DOE’s Office of Science Environmental Systems Science (ESS) program (see Additional files, Supplemental Table 4).
Prior to sample registration, we discussed expected sample types involved, how to assign IGSNs and link related samples, essential metadata needed to understand specific sample types, and past sample tracking workflows with representatives from each project. Some projects had already collected samples and preferred to register for IGSNs after collection to be associated with digital files, while other projects pre-registered their samples before collection, or registered directly after collection. We used initial feedback and background research to identify several core descriptive sample metadata fields likely to be necessary for searches on ESS-DIVE to be most effective, including standardized information on the following[45] (also see Additional files, Supplemental Table 3 for the full translation table comparing metadata elements from existing standards and templates):
- IGSN and Parent IGSN (where relevant)
- Sample Name (project-specific sample name, must be unique)
- Chief Scientist/Collector
- Sample Type fields:
- Object Type (e.g. Individual sample, core, site),
- Material (e.g. Liquid-aqueous, Rock, Soil, Biology),
- Sampled Feature (primary physiographic feature sample collected from)
- Location Information (Latitude, Longitude in WGS84; Location description),
- Date (ISO 8601; e.g., 1954-04-07),
- Collection Method Description
- Project
Note that this list represents the initial IGSN metadata fields that should be required and were subsequently revised after our pilot test work. Many additional metadata fields are available and are recommended or optional depending on the sample type.[12]
The researchers involved in our testing used SESAR’s sample management portal MySESAR to register samples and update metadata. We recommended a specific workflow for participants to register their samples and update sample collection metadata, outlined in our GitHub repository[46] and associated dataset.[45]
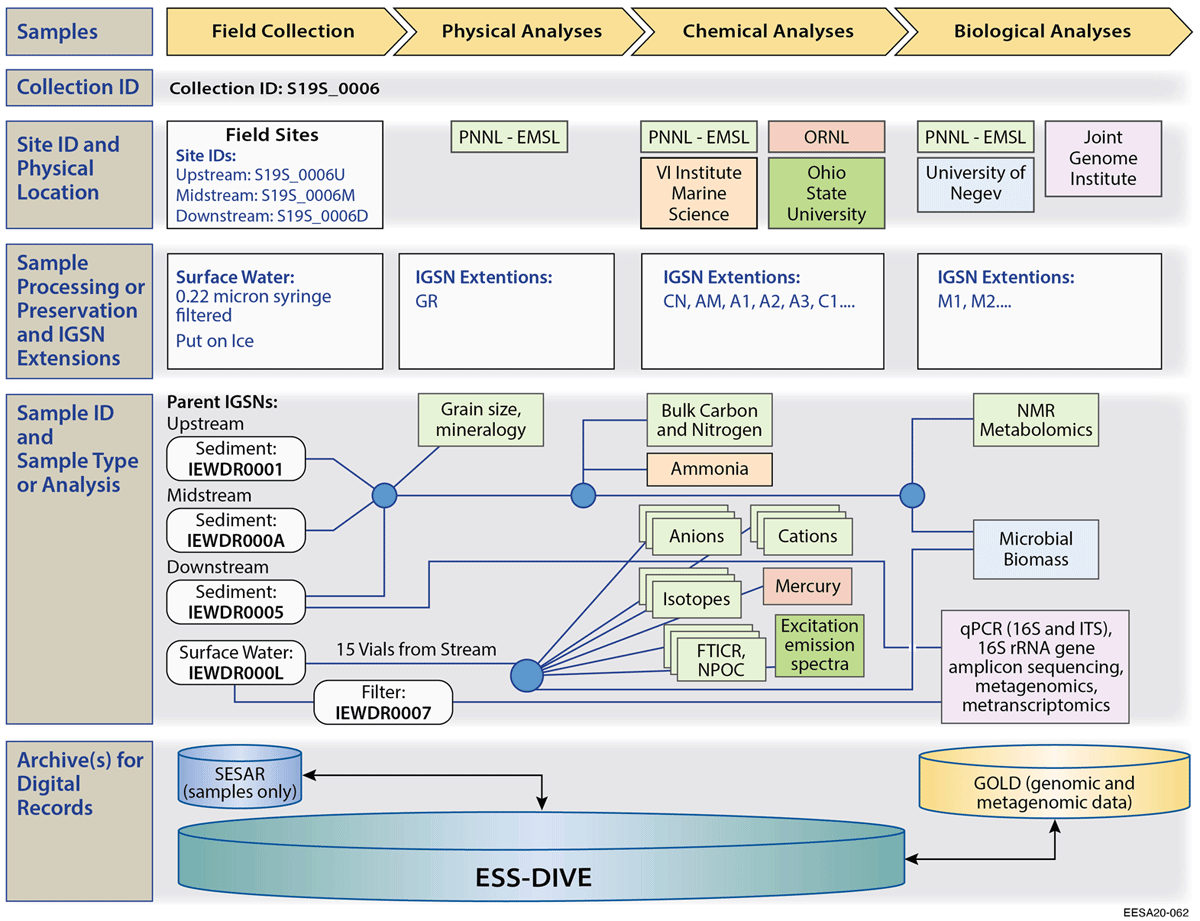
We also worked with individuals to map sample history from collection of samples in the field through a variety of analyses, and publication (Figure 2). This exercise helped us determine sample tracking needs and develop recommendations for assigning PIDs and linking highly-related samples and subsamples.
|
After sample collection and registration, we asked and discussed the following questions:
- What sample collection metadata is needed to understand resulting sample data?
- How much effort did it take to register samples and standardize metadata?
- What is needed to make sample PID registration and standardization easier?
Developing the final IGSN-ESS reporting guidelines
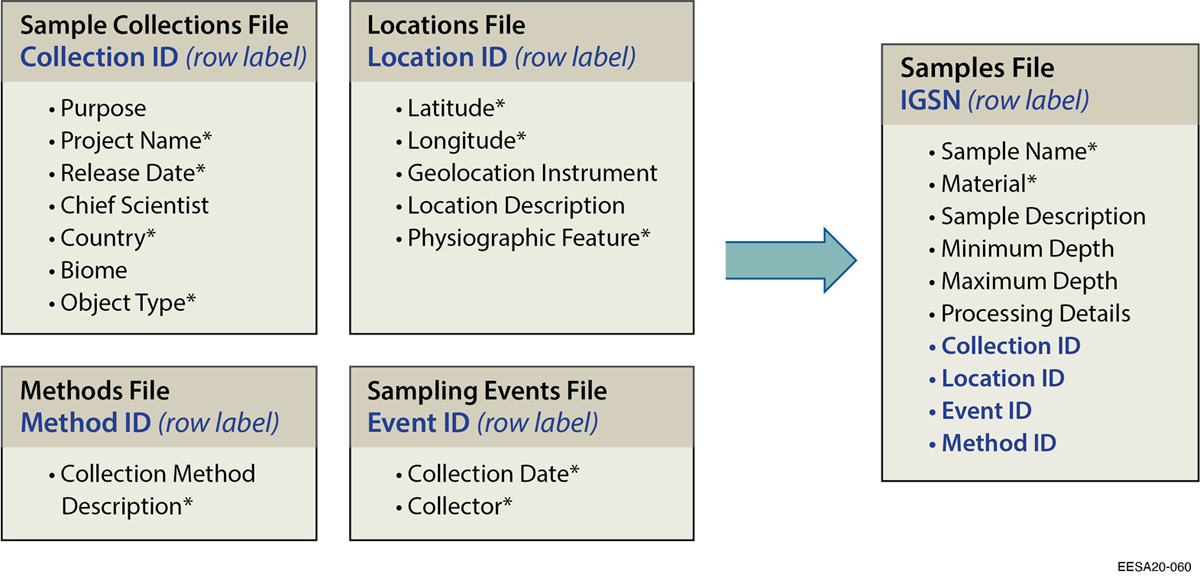
We used a combination of research on existing standards and pilot test feedback to develop final recommendations for allocating identifiers and assigning standard metadata.[45] We took extensive notes during meetings with pilot test participants, and we compiled specific feedback on improving guidance on allocating identifiers and relationships, metadata needed to understand relevant sample types, and improve efficiency of sample registration and standardization. Pilot test participants identified metadata elements that needed to be added, modified, or removed to improve relevance for multidisciplinary ecosystem science samples. We then used our translation table (see Additional files, Supplemental Table 3) comparing other existing standards to guide-specific recommendations. For example, to address feedback regarding inefficiencies in providing all metadata at individual sample levels, we added the Darwin Core elements Location ID, Collection ID, and Event ID. We then reviewed existing, commonly-used ontologies (ENVO, BCO, PO) to select important vocabulary terms to characterize sample type, material, and environmental context. We developed a list of relevant terms based on pilot test studies, and all participants helped decide on our final term lists for object type and material, specifically.
All feedback was addressed in our final recommendations, which we compiled into GitHub, and more user-friendly GitBook documentation. This documentation includes instructions on registering samples for IGSNs using our revised template, specific definitions/instructions/examples for each metadata element, lists of terms for elements where controlled vocabulary is needed, and instructions for how to contribute feedback using GitHub, and how to cite the final format. To develop documentation, we used the ESS-DIVE community GitHub for samples, inspired from user-friendly documentation for Darwin Core, which facilitates additional community feedback (through public GitHub issues) and versioning. We presented our final recommendations and documentation in two additional community webinars, which are advertised to ESS-DIVE users and ESS scientists, and published on the ESS-DIVE website. The purpose of community webinars was to present our conclusions and collect any additional feedback.
As a community-oriented data repository, we will continue to gather feedback and develop additional tools to support users in submitting, searching for, integrating, and reusing high-quality sample data.
Results
Review of existing sample identifier and metadata practices
In our review, we found that numerous studies have documented that persistent identifiers (PIDs) enable sample tracking across facilities and publications, and support reuse over time.[2][24][28][48][49][50][51][52][53] PIDs are globally unique, stored with descriptive metadata, and arguably essential for supporting data synthesis.[50][51] While there are several options for obtaining PIDs—Archival Resource Keys (ARKs), Digital Object Identifiers (DOI), and Uniform Resource Identifier (URI)[28][50][51][52][54]—the International GeoSample Number (IGSN) is the primary PID for physical samples (Table 1 and Additional files, Supplemental Table 1).[51][55][56] IGSNs were originally designed for geoscience samples, but they have been used for a variety of biological and environmental sample types. The IGSN organization is now expanding to better support multidisciplinary samples, and it acts as lead of the Internet of Samples project.[57]
Through community discussions, we determined that the most important factors in selecting a PID were an international community with expertise on sample documentation; associated sample-specific metadata that will eventually enable global sample search and integration; and user-friendly infrastructure to mint PIDs, validate metadata, and provide a sample-specific web landing page (Additional files, Supplemental Table 1). IGSNs are the only identifier with these characteristics, as they are uniquely governed by an international community organization (IGSN e.V.), with a mission to mint and maintain persistent identifiers for physical samples. SESAR is the largest IGSN allocating agent, which has enabled us to readily test the process of sample registration and standardizing metadata without first building new infrastructure to mint PIDs, print IGSN barcode labels, and submit and validate metadata. SESAR also provides a persistent sample landing page (e.g., IGSN:IEBWE000L) with metadata and links to related resources.[2][48][51][58][59]
Through our comparison of metadata elements in existing sample-related standards and templates (Additional files, Supplemental Table 3), we concluded that IGSN metadata contains basic information needed, and was therefore sufficient to use in our pilot for standardizing sample metadata.
Sample identifier and metadata testing in the field
Our pilot test included eight DOE ESS-supported projects that collected field-based samples, including studies of biogeochemical responses to contamination, climate change, or other disturbances (Additional files, Supplemental Table 4). Project sample types included soil cores, core sections, individual soil samples, sediment, gas, porewater, pond water, river water, leaves, and biofilms. Researchers registered their samples with IGSNs to determine practicalities of using the original SESAR IGSN template (i.e., an Excel spreadsheet with sample metadata elements for each column and unique sample names/IGSNs for each row)[12] in multidisciplinary scientific workflows.
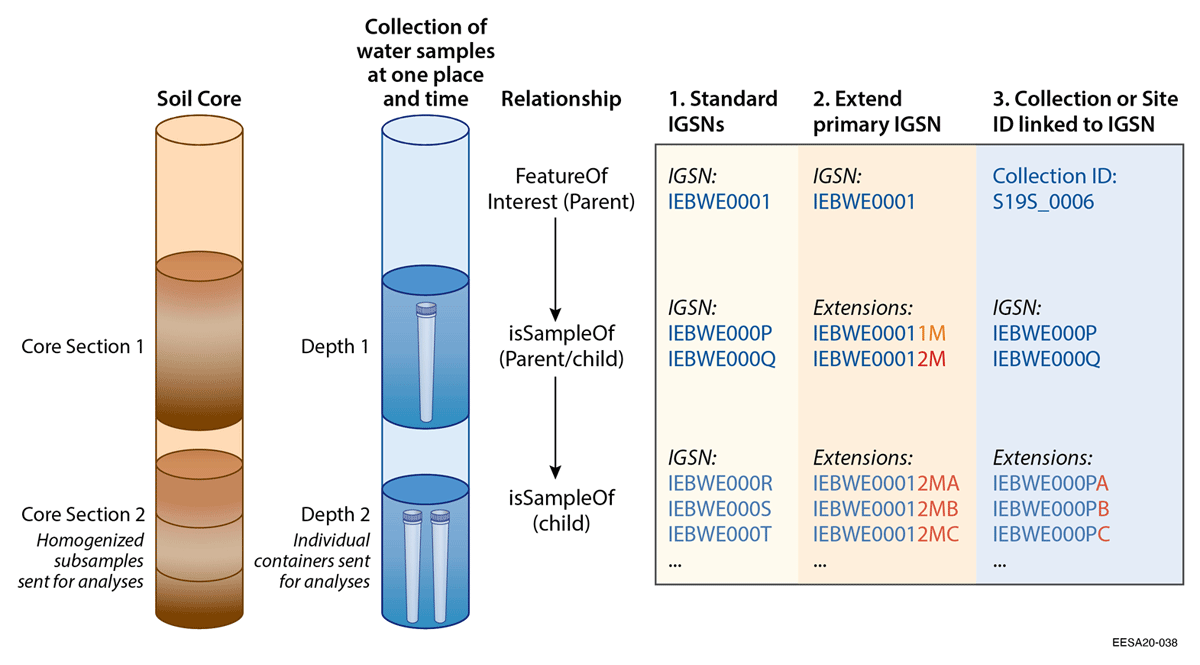
A total of 4,485 IGSNs were registered as part of the pilot (Additional files, Supplemental Table 4). A primary sample for participating projects was often split into multiple subsamples or replicates and sent to different labs (2–9 labs/user facilities) for numerous analyses (2–23 analyses) (Figure 2 and Additional files, Supplemental Table 4).[17][18][20][47] There was universal agreement among researchers that top-level “parent” samples (e.g., soil core), and related “child” samples (e.g., subsections of a soil core) be assigned individual IGSNs. Note that a soil core is a physical parent sample, while in some cases researchers may need to link a set of related samples with no physical parent sample. One example from our test was a set of water samples collected at different depths at a specific point and time in a pond. (Figure 3)
|
Most participants were uncertain whether to assign new IGSNs to subsamples or replicates stored in different containers or split for analyses, particularly when they are essentially considered to be the same sample with the same metadata; many researchers preferred qualifiers/extensions from the same primary IGSN in such cases (Figure 3).[24] IGSN extensions are currently allowed by request through SESAR IGSN, and are preferred by some users to avoid numerous rounds of IGSN registration and redundant metadata entry. The extensions can allow precise provenance tracking and incorporate additional analytical metadata when subsamples are sent out for a variety of analyses, without requesting new IGSNs. However, this requires users to ensure that their extensions are unique and are restricted to a limited number of additional characters, and that they are batch registered through the IGSN allocating agent with associated metadata, including at least object/sample type, sample name, and the parentIGSN (and ideally all relevant metadata inherited from the parentIGSN). IGSN allocating agents could consider more efficient approaches for registering IGSN subsamples with the same metadata as parentIGSNs, such as adding a metadata field to list subsamples (IGSNs with user-specified extensions), or to have extended IGSNs automatically resolve to the primary IGSN landing page, as done by the ARK identifier system for containment qualifiers.[60]
Researchers also had different opinions on whether related entities (e.g., location) should get an IGSN/PID. In most cases, project-specific, locally unique IDs were sufficient for collection and location IDs. Some researchers assigned IGSNs to wells that were re-sampled over time.
Use of IGSN metadata and template
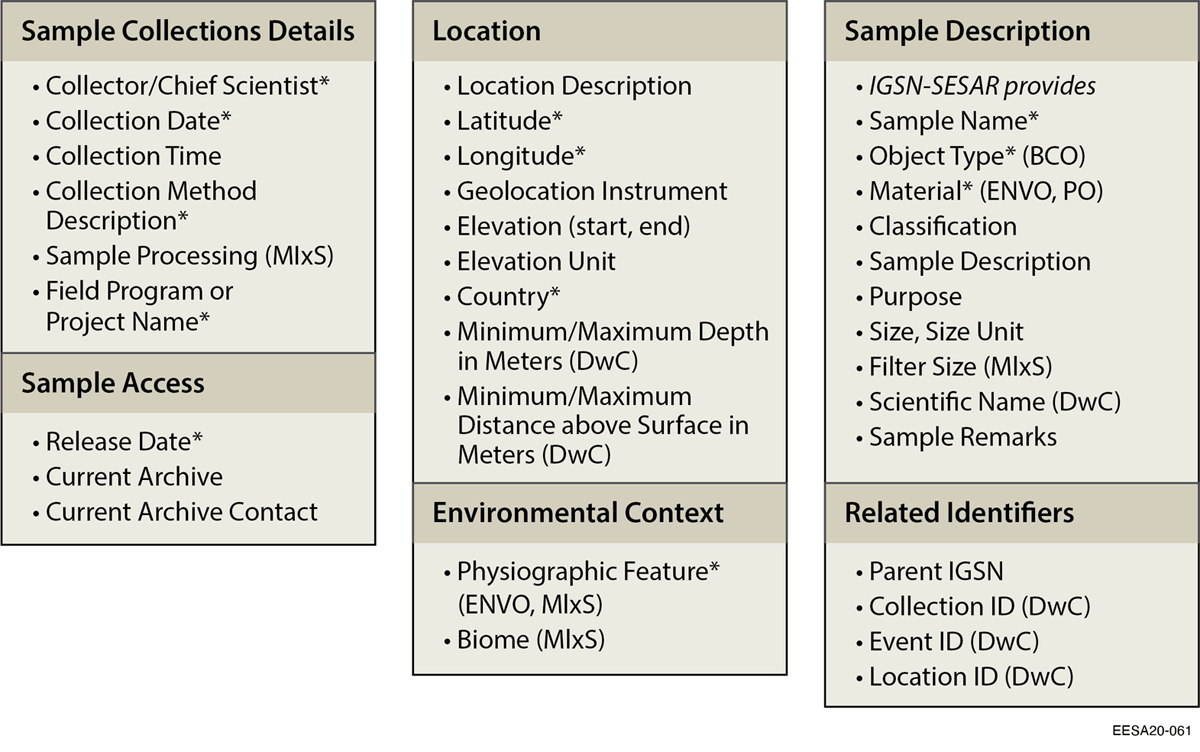
Much of the IGSN Core Descriptive Metadata is relevant for samples across research domains, but there are key metadata fields and vocabulary terms that are missing or do not accurately describe some ecological samples. We added two essential metadata elements from Minimum Information about any Sequence (MIxS) (i.e., broad environmental context/biome, sample processing)[10], and added or modified fields based on DarwinCore (i.e., "Scientific Name," "Depth," and "Height" fields) to more fully describe ecosystem samples (Figure 5). We concluded that the Environment Ontology (ENVO) includes more relevant terms to describe sample material and environmental context for ecosystem science samples. Because ENVO is used in the MIxS template, it also helps improve interoperability when relating geoscience analyses with omics analyses for samples (Table 2), which is often important in ecosystem studies.
| ||||||||||||||||||
IGSN was designed to allow community-specific metadata profiles along with common high-level metadata to support broader interoperability. However, variations across the communities in high-level vocabularies, such as object/sample type and material terms, can inhibit interoperability if the vocabulary terms are not well defined, managed, and linked. We therefore mapped SESAR IGSN terms to ENVO terms for materials. Unlike IGSN vocabulary terms, ENVO terms have specified definitions, PIDs, and are linked to other related terms across many existing ontologies. We also believe that the broader IGSN community could contribute valuable input to the ENVO terms, and benefit from using this ontology or others as they move towards supporting a wider variety of disciplines. We found community agreement that the IGSN Object type terms also need to be revised, and high-level vocabularies will be addressed in the new ESIP Physical Sample Curation cluster.[61]
Participants with extensive sampling campaigns found that the spreadsheet format requiring full documentation for each individual sample was impractical. To partially address this, we follow DarwinCore by adding the option of managing metadata using identifiers for higher-level entities (collectionID, locationID, eventID) to help avoid redundant metadata entry. Managing metadata for larger collections of samples by describing sample collections, locations, or events in separate files (see Figure 4) can allow programmatic transfer of relevant metadata to individual samples. However, with regards to applying IGSN metadata to locations, we encountered several issues (described in Table 3) as metadata was not intended to fully document site information. We provide additional recommendations in Box 1 that may further improve the efficiency of standardizing sample metadata and/or address practical concerns of researchers.
|
| ||||||||||||||
| Box 1. Efficiency recommendations for large sampling campaigns |
| 1. Field collection apps: On- or off-line field collection apps can be programmed with standard metadata and enable users to collect information directly in the field, such as automated generation of date/time, and location.[3][62] Apps could also generate and record PIDs in the field, and be paired with portable label printers. 2. Label material: For IGSNs to be associated with the physical sample, recommended label material and adhesive to withstand extreme conditions (e.g., –80 freezer, water submersion) is useful. Some specific recommendations include: waterproof or cryogenic labels (e.g., via LabTAG), vinyl or polyester labels (e.g., via DYMO), and Tough-Tags (via Diversified Biotech). 3. Barcodes and APIs: Sample label barcodes and barcode readers could utilize an API for effectively pulling specific metadata from the IGSN record (e.g., sample type, location) to assist with downstream data analysis or processing, and/or automatically adding links to additional metadata or data as it is produced later in the lifecycle of a sample.[63] |
Sample identifiers for tracking and linking
Researchers generally use their own meaningful sample name for internal sample tracking and individual data analysis workflows (Figure 2); so, both the project-specific sample name and the IGSN should be associated with digital records of the sample. The IGSN, as a globally unique PID, is better suited for automated sample tracking and linking related information over the data life cycle, from field-collection to open-access publication. (Guralnick et al, 2014; Lehnert et al, 2019a) With IGSNs, related samples can be more clearly linked on the sample landing page (e.g., IGSN:IEWDR000X). Further, specific location or event IDs clarify common relationships for samples and derivatives in a project studying ecological processes at a given location, e.g., involving plant litter, leaf, root, soil, and associated omics samples.
To most effectively link samples, we recommend that all labs and data systems that generate or store sample data utilize the IGSN or other PID, adding it to metadata templates where relevant. Use of the SESAR API to obtain relevant information about samples can facilitate reuse of metadata across multiple labs or facilities. In theory, the IGSN could be used to automatically add links on the sample landing page to data generated at different facilities; however, no tools are currently available to enable automated linkages.
Improvements are needed to link environmental and associated biological samples. Genomic samples, for example, should be assigned a BioSample number when submitted for sequencing, and linked to the original field-collected sample where relevant (Table 2). There is currently no automated way to link such identifiers, so we recommend providing a full link of the IGSN landing page in the source material ID field in the MIxS template (Table 2).
Discussion
Sample PIDs and metadata in multidisciplinary environmental sciences
We advocate use of IGSNs for ecosystem science samples for a number of reasons. IGSNs are the only PID specifically designed for samples with associated metadata.[51] IGSN is the only PID backed by an international community of experts, dedicated to identifying, describing, and linking sample data.[64] Participation in the IGSN community will help improve the usefulness of sample PIDs and relevance of associated metadata for multidisciplinary ecosystems science. Additionally, other large national agencies have or plan to adopt IGSNs (e.g., United States Geological Survey [USGS], National Oceanic and Atmospheric Administration [NOAA], Commonwealth Scientific and Industrial Research Organisation [CSIRO]). A recently funded effort, iSamples, will improve infrastructure for samples that utilize IGSN and other sample PIDs, and eventually support global search for an even wider variety of sample types.[57]
Benefits to data contributors and users
Funders of scientific research, such as the U.S. DOE and the National Science Foundation (NSF), require robust data management and publication plans, which should include details for managing and tracking valuable sample data. These data are often not well-described and are missing key information needed to interpret and reuse it, leading to data loss.[13][65][66] The IGSN-ESS reporting format can assist ecosystem researchers in creating effective sample management plans and preserving their data.
More widespread use of sample PIDs and related metadata will help make sample data more FAIR.[26][51] Standard information to characterize the sample type, location, and date are particularly useful for finding relevant data.[3][62] Persistent landing pages for samples allow long-term access to sample data and metadata. Use of a controlled vocabulary for key metadata (e.g., sample material and environmental context) helps make data interoperable and more easily integrated across datasets. In addition, reuse often requires information on collection and processing methods.[62] Samples with standard metadata can be more easily shared (i.e., understood and reused) with collaborators, which helps avoid situations where information is lost when people change institutions or retire.[67] High-quality published data increasingly helps scientists achieve greater academic recognition, higher citation rates, and can lead to new opportunities for co-authorship and collaboration.[68][69]
Multidisciplinary ecosystem science often involves complex workflows, and sample PIDs and common metadata provide essential information to help users automatically track samples and add relevant data throughout the sample life cycle. PIDs (such as IGSNs and DOIs) are essential for tracking use of samples and related data over time.[51][52][63] This provides the foundation to build tools that automatically link and exchange this information across data systems, with no further input from the user after the initial metadata is provided.
Ecosystems research often relies on sample data combined with other data types, such as remote sensing and environmental sensor data, to answer questions about ecosystem response to increasingly rapid global changes.[18][19][22][70] One limitation is that our standards comparison was focused on sample-related metadata; we need more work towards incorporating standards suitable for other related entities, such as locations and sensors.[31][71][72]
More widespread standardization will help reduce the estimated 80% effort currently spent on data wrangling for synthesis work, and enable more efficient data integration and analysis.[67] Improved sample data management and reuse will increase the pace of scientific discovery and accelerate new fields of enquiry.[13][67] Already, publicly available nucleic acid sequences have enabled scientists to build phylogenies and perform comparative genomics studies, and are now essential in community ecology.[73] Biodiversity records are regularly combined with climate and land use data to predict species distributions, biodiversity, and explore multi-scale ecological patterns.[39][67][74][75] With our multidisciplinary reporting format, we can move beyond infrastructure supporting individual data types, towards efficiently integrating multidisciplinary data to understand ecosystem processes from molecular to global scales.
Conclusions
Summary of IGSN-ESS identifier and metadata recommendations
Many multidisciplinary projects have complicated workflows and need an efficient system for tracking samples as they are sent to different collaborators, labs, user facilities, and published online (Figure 1). Despite growing need and interest, there was previously no straightforward guidance on how to describe sample collections or multidisciplinary samples. Based on our work, we therefore recommend registering samples with IGSNs, using IGSN-ESS's modified metadata template for ecosystem sciences (Figure 5). The downloadable template, along with complete definitions of all terms and instructions for IGSN registration using IGSN-ESS and how to provide feedback are detailed in the ESS-DIVE community GitHub repository, as well as an associated data publication.[45]
|
To avoid redundancy in describing samples with the same metadata, we add the optional practice of assigning common sample metadata to a collection, location, or event.[11][76][77][78] A collection ID provides a flexible way for projects to define common metadata for any set of related samples, while location ID can be used to describe project locations/sites, and event ID can describe metadata for a given sampling event (see Figure 4). These related IDs also provide an unambiguous way to automatically link commonly-related samples. This is particularly important for ecosystem science research, as diverse sample types often need to be clearly linked by specific related identifiers (e.g., location).
For highly-related subsamples with the same metadata, we recommend the option of ID extensions, which could be opaque or meaningful as long as they are unique (Figure 3). It would further improve efficiency of subsample IGSN registration to update the primary IGSN metadata by listing the subsamples or replicates under a “subsample” field, instead of registering them separately. Or the IGSN resolution service could follow the practice of ARKs, where IGSNs with extensions (i.e., containment qualifiers) (see Additional files, Supplemental Table 1) automatically resolve to the primary IGSN landing page.
We added or revised fields and vocabulary terms to more accurately describe multidisciplinary samples, and support data linking and reusability (Figure 5). We include controlled vocabularies for relevant subsets of terms from ENVO[42][45], which improves description, search, and integration of a variety of multidisciplinary sample types using key fields (e.g., sample type, sample material, and environmental context).[45] We selected terms based on an evaluation of their relevance and likelihood of being used in multiple contexts. We also found that use of ENVO for both local (physiographic feature) and broad (biome) environmental context (e.g., stream ENVO_00000023) is important to fully characterize soil, sediment, and water samples.
Promoting adoption and other next steps
Most ecologists and environmental scientists now understand the importance of data archiving, but struggle to manage data effectively.[13][67] Removing even trivial barriers can increase the likelihood that researchers will adopt beneficial practices that take effort to achieve.[79] User-friendly guidance and sample metadata templates are an essential step in promoting standard practices that make data publishing, integration, and reuse easier. However, investments are also needed in training programs[80], tools to assist with legacy data and analytical instrument systems, and improved data quality management systems that encourage good management practices throughout the research process.[81][82] We need tools that translate across existing metadata conventions and use sample and relationship metadata to automatically generate digital resource maps; this could promote adoption by helping users precisely document sample history and linkages to other PIDs and documents.[72][83] Global sample search (e.g., iSamples Central)[57], with integrated results, based on key fields (e.g., sample material, location, environmental context, methods, and associated data variables/analyses) would greatly enhance sample data discovery and reuse, and is likely the most effective tool to promote widespread adoption of sample standards (e.g., GBIF).[38]
Overcoming complex challenges that require communities to change behavior and provide standardized data will require a coordinated effort, which is best addressed by collaborations of key stakeholders who establish community consensus, enforce guidelines, and help solve problems.[82][84] These stakeholders include a variety of data contributors and users from different scientific domains, as well as laboratory facilities, repositories, funders, and publishers that take part in institutionalizing and rewarding good data management practices[82][85][86][87] Community coordination on sample reporting conventions and linked cyberinfrastructure will help solve data management problems, expand access pathways, and make our sample data more useful over time.
Additional files
The additional files for this article can be found as follows:
- Supplemental Table 1: Comparison of ARK and IGSN sample identifiers characteristics
- Supplemental Table 2: Overview of existing sample-related standards and templates
- Supplemental Table 3: Translation table comparing standards and templates related to sample metadata
- Supplemental Table 4: Summary of projects involved in IGSN and standard metadata pilot test
Acknowledgements
We greatly appreciate access to SESAR IGSN infrastructure, which allowed us to test use of IGSNs and standardized descriptive metadata with the DOE ESS community. We especially thank Kerstin Lehnert and Sarah Ramdeen for help with sample registration through SESAR, guidance with sample metadata, and for their work with organizing community workshops and meetings regarding samples. JED participated in the IGSN 2040 Steering Committee, and thanks all committee members who contributed to ideas on IGSN value propositions, business planning, and insight regarding direction of IGSN. We also thank Jens Klump for guidance on related work, Chris Mungall for guidance on ontology use and ENVO. We thank other members of the ESS-DIVE team (including Shreyas Cholia, Karen Whitenack) and the National Center for Ecological Analysis and Synthesis (NCEAS) and DataONE (Matt Jones and Chris Jones) who provided information on metadata standards and identifier use across the DataONE network. And, we thank all those who contributed to a variety of community discussions on sample identifiers within the U.S. DOE Biological and Environmental Research community, and broader groups and discussions across ESIP, RDA, and USGS. Thanks also to Diana Swantek for assistance with final figure designs.
Author contributions
JED, CV, and DA conceived the project. JED conducted the metadata review, led the pilot test, synthesized results and wrote the manuscript. CV and DA supervised the project and provided guidance on its execution. JED, MB, TH, ER, and RW contributed to the comparison of existing sample identifiers, metadata standards and/or reporting conventions. KB, MB, KDC, RJEA, KSE, AEG, VH, ZK, NM, ZP, ER, PS, JCS, PW, MZ participated in field testing and provided feedback on identifiers and metadata. JED and RCO created github documentation. MB, RCO, HE, VH provided input into the reporting format development and documentation as members of the ESS-DIVE repository team. All authors contributed to discussions on the project, as well as reviewing and editing the manuscript.
Funding information
ED, CV, MB, RCO, HE, VH, and DA were funded through the ESS-DIVE repository by the U.S. DOE’s Office of Science Biological and Environmental Research under contract number DE-AC02-05CH11231 to LBNL, as part of its Earth and Environmental Systems Science Division Data Management program. KSE was supported by the United States Department of Energy contract No. DE-SC0012704 to Brookhaven National Laboratory. RJEA was supported by the U.S. Department of Energy Office of Science, Office of Biological and Environmental Research under Contract No. DE-AC02-05CH11231 to LBNL, as part of the Terrestrial Ecosystem Science Program. ELB, PS, and ZK contributions were supported as part of the Watershed Function Scientific Focus Area funded by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under Award Number DE-AC02-05CH11231. AEG and JCS were supported by the U.S. DOE-BER, as part of BER’s Subsurface Biogeochemistry Research (SBR) Program at Pacific Northwest National Laboratory (PNNL), which is operated by Battelle Memorial Institute for the U.S. DOE under Contract No. DE-AC05-76RL01830. ABK, NM, and MZ were supported by the Department of Energy, Office of Science, Biological and Environmental Research, Subsurface Biogeochemical Research program (SCW1053) and performed under the auspices of the U.S. Department of Energy by Lawrence Livermore National Laboratory under Contract DE-AC52-07NA27344. RLW’s contribution was supported by a National Science Foundation grant 2004562.
Data accessibility statement
Data and recommended metadata guidelines generated as part of this work are published in the ESS-DIVE repository and the the work of Damerow et al. Future updates will be managed and available through our community GitHub repository.
Competing interests
The authors have no competing interests to declare.
References
- ↑ Weart, Spencer (26 February 2013). "Rise of interdisciplinary research on climate". Proceedings of the National Academy of Sciences 110 (Supplement 1): 3657–3664. doi:10.1073/pnas.1107482109. PMC PMC3586608. PMID 22778431. https://www.pnas.org/content/110/Supplement_1/3657.
- ↑ 2.0 2.1 2.2 2.3 Devaraju, A.; Klump, J.; Cox, S.J.D. et al. (1 November 2016). "Representing and publishing physical sample descriptions" (in en). Computers & Geosciences 96: 1–10. doi:10.1016/j.cageo.2016.07.018. ISSN 0098-3004. https://www.sciencedirect.com/science/article/pii/S0098300416302023.
- ↑ 3.0 3.1 3.2 Ponsero, Alise J; Bomhoff, Matthew; Blumberg, Kai; Youens-Clark, Ken; Herz, Nina M; Wood-Charlson, Elisha M; Delong, Edward F; Hurwitz, Bonnie L (31 July 2020). "Planet Microbe: a platform for marine microbiology to discover and analyze interconnected ‘omics and environmental data". Nucleic Acids Research 49 (D1): D792–D802. doi:10.1093/nar/gkaa637. ISSN 0305-1048. PMC PMC7778950. PMID 32735679. https://academic.oup.com/nar/article/49/D1/D792/5879428.
- ↑ Cordeiro, Amanda L.; Norby, Richard J.; Andersen, Kelly M.; Valverde-Barrantes, Oscar; Fuchslueger, Lucia; Oblitas, Erick; Hartley, Iain P.; Iversen, Colleen M. et al. (2020). "Fine-root dynamics vary with soil depth and precipitation in a low-nutrient tropical forest in the Central Amazonia" (in en). Plant-Environment Interactions 1 (1): 3–16. doi:10.1002/pei3.10010. ISSN 2575-6265. https://onlinelibrary.wiley.com/doi/abs/10.1002/pei3.10010.
- ↑ Malik, Ashish A.; Martiny, Jennifer B. H.; Brodie, Eoin L.; Martiny, Adam C.; Treseder, Kathleen K.; Allison, Steven D. (1 January 2020). "Defining trait-based microbial strategies with consequences for soil carbon cycling under climate change" (in en). The ISME Journal 14 (1): 1–9. doi:10.1038/s41396-019-0510-0. ISSN 1751-7370. PMC PMC6908601. PMID 31554911. https://www.nature.com/articles/s41396-019-0510-0.
- ↑ Treseder, Kathleen K.; Balser, Teri C.; Bradford, Mark A.; Brodie, Eoin L.; Dubinsky, Eric A.; Eviner, Valerie T.; Hofmockel, Kirsten S.; Lennon, Jay T. et al. (3 September 2011). "Integrating microbial ecology into ecosystem models: challenges and priorities". Biogeochemistry 109 (1-3): 7–18. doi:10.1007/s10533-011-9636-5. ISSN 0168-2563. http://dx.doi.org/10.1007/s10533-011-9636-5.
- ↑ Soranno, Patricia A.; Schimel, David S. (2014). "Macrosystems ecology: big data, big ecology" (in en). Frontiers in Ecology and the Environment 12 (1): 3–3. doi:10.1890/1540-9295-12.1.3. ISSN 1540-9309. https://onlinelibrary.wiley.com/doi/abs/10.1890/1540-9295-12.1.3.
- ↑ 8.0 8.1 8.2 Field, Dawn; Amaral-Zettler, Linda; Cochrane, Guy; Cole, James R.; Dawyndt, Peter; Garrity, George M.; Gilbert, Jack; Glöckner, Frank Oliver et al. (21 June 2011). "The Genomic Standards Consortium" (in en). PLOS Biology 9 (6): e1001088. doi:10.1371/journal.pbio.1001088. ISSN 1545-7885. PMC PMC3119656. PMID 21713030. https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.1001088.
- ↑ Reddy, T.B.K.; Thomas, Alex D.; Stamatis, Dimitri; Bertsch, Jon; Isbandi, Michelle; Jansson, Jakob; Mallajosyula, Jyothi; Pagani, Ioanna et al. (27 October 2014). "The Genomes OnLine Database (GOLD) v.5: a metadata management system based on a four level (meta)genome project classification". Nucleic Acids Research 43 (D1): D1099–D1106. doi:10.1093/nar/gku950. ISSN 1362-4962. PMC PMC4384021. PMID 25348402. https://academic.oup.com/nar/article/43/D1/D1099/2439522.
- ↑ 10.0 10.1 10.2 10.3 Yilmaz, Pelin; Kottmann, Renzo; Field, Dawn; Knight, Rob; Cole, James R.; Amaral-Zettler, Linda; Gilbert, Jack A.; Karsch-Mizrachi, Ilene et al. (1 May 2011). "Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications" (in en). Nature Biotechnology 29 (5): 415–420. doi:10.1038/nbt.1823. ISSN 1546-1696. PMC PMC3367316. PMID 21552244. https://www.nature.com/articles/nbt.1823.
- ↑ 11.0 11.1 11.2 11.3 Wieczorek, John; Bloom, David; Guralnick, Robert; Blum, Stan; Döring, Markus; Giovanni, Renato; Robertson, Tim; Vieglais, David (6 January 2012). "Darwin Core: An Evolving Community-Developed Biodiversity Data Standard" (in en). PLOS ONE 7 (1): e29715. doi:10.1371/journal.pone.0029715. ISSN 1932-6203. PMC PMC3253084. PMID 22238640. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0029715.
- ↑ 12.0 12.1 12.2 12.3 System For Earth Sample Registration (SESAR) (6 February 2020) (in en). SESAR Batch Registration Quick Guide. doi:10.5281/ZENODO.3874923. https://zenodo.org/record/3874923.
- ↑ 13.0 13.1 13.2 13.3 Roche, Dominique G.; Kruuk, Loeske E. B.; Lanfear, Robert; Binning, Sandra A. (10 November 2015). "Public Data Archiving in Ecology and Evolution: How Well Are We Doing?" (in en). PLOS Biology 13 (11): e1002295. doi:10.1371/journal.pbio.1002295. ISSN 1545-7885. PMC PMC4640582. PMID 26556502. https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.1002295.
- ↑ Treloar, Andrew; Klump, Jens (20 December 2019). "Updating the Data Curation Continuum" (in en). International Journal of Digital Curation 14 (1): 87–101. doi:10.2218/ijdc.v14i1.643. ISSN 1746-8256. http://www.ijdc.net/article/view/643.
- ↑ Chase, John H.; Bolyen, Evan; Rideout, Jai Ram; Caporaso, J. Gregory (22 December 2015). "cual-id: Globally Unique, Correctable, and Human-Friendly Sample Identifiers for Comparative Omics Studies" (in EN). mSystems. doi:10.1128/mSystems.00010-15. PMC PMC5069752. PMID 27822516. https://journals.asm.org/doi/abs/10.1128/mSystems.00010-15.
- ↑ Varadharajan, C.; Cholia, S.; Snavely, C. et al. (8 January 2019). "Launching an Accessible Archive of Environmental Data" (in en-US). Eos. doi:10.1029/2019eo111263. http://eos.org/science-updates/launching-an-accessible-archive-of-environmental-data.
- ↑ 17.0 17.1 Biological and Environmental Research Advisory Committee (2017). "Grand Challenges for Biological and Environmental Research: Progress and Future Vision" (PDF). U.S. Department of Energy. https://genomicscience.energy.gov/BERfiles/BERAC-2017-Grand-Challenges-Report.pdf.
- ↑ 18.0 18.1 18.2 Chadwick, K. Dana; Brodrick, Philip G.; Grant, Kathleen; Goulden, Tristan; Henderson, Amanda; Falco, Nicola; Wainwright, Haruko; Williams, Kenneth H. et al. (2020). "Integrating airborne remote sensing and field campaigns for ecology and Earth system science" (in en). Methods in Ecology and Evolution 11 (11): 1492–1508. doi:10.1111/2041-210X.13463. ISSN 2041-210X. https://onlinelibrary.wiley.com/doi/abs/10.1111/2041-210X.13463.
- ↑ 19.0 19.1 Serbin, Shawn P.; Wu, Jin; Ely, Kim S.; Kruger, Eric L.; Townsend, Philip A.; Meng, Ran; Wolfe, Brett T.; Chlus, Adam et al. (2019). "From the Arctic to the tropics: multibiome prediction of leaf mass per area using leaf reflectance" (in en). New Phytologist 224 (4): 1557–1568. doi:10.1111/nph.16123. ISSN 1469-8137. https://onlinelibrary.wiley.com/doi/abs/10.1111/nph.16123.
- ↑ 20.0 20.1 20.2 Stegen, James C.; Goldman, Amy E. (9 October 2018). "WHONDRS: a Community Resource for Studying Dynamic River Corridors" (in EN). mSystems. doi:10.1128/mSystems.00151-18. PMC PMC6178584. PMID 30320221. https://journals.asm.org/doi/abs/10.1128/mSystems.00151-18.
- ↑ Wu, Jin; Rogers, Alistair; Albert, Loren P.; Ely, Kim; Prohaska, Neill; Wolfe, Brett T.; Oliveira, Raimundo Cosme; Saleska, Scott R. et al. (2019). "Leaf reflectance spectroscopy captures variation in carboxylation capacity across species, canopy environment and leaf age in lowland moist tropical forests" (in en). New Phytologist 224 (2): 663–674. doi:10.1111/nph.16029. ISSN 1469-8137. https://onlinelibrary.wiley.com/doi/abs/10.1111/nph.16029.
- ↑ 22.0 22.1 Wu, Jin; Serbin, Shawn P.; Ely, Kim S.; Wolfe, Brett T.; Dickman, L. Turin; Grossiord, Charlotte; Michaletz, Sean T.; Collins, Adam D. et al. (2020). "The response of stomatal conductance to seasonal drought in tropical forests" (in en). Global Change Biology 26 (2): 823–839. doi:10.1111/gcb.14820. ISSN 1365-2486. https://onlinelibrary.wiley.com/doi/abs/10.1111/gcb.14820.
- ↑ Beck, Marcus W.; O’Hara, Casey; Lowndes, Julia S. Stewart; Mazor, Raphael D.; Theroux, Susanna; Gillett, David J.; Lane, Belize; Gearheart, Gregory (20 July 2020). "The importance of open science for biological assessment of aquatic environments" (in en). PeerJ 8: e9539. doi:10.7717/peerj.9539. ISSN 2167-8359. PMC PMC7377246. PMID 32742805. https://peerj.com/articles/9539.
- ↑ 24.0 24.1 24.2 Conze, Ronald; Lorenz, Henning; Ulbricht, Damian; Elger, Kirsten; Gorgas, Thomas (25 January 2017). "Utilizing the International Geo Sample Number Concept in Continental Scientific Drilling During ICDP Expedition COSC-1" (in en). Data Science Journal 16: 2. doi:10.5334/dsj-2017-002. ISSN 1683-1470. http://datascience.codata.org/articles/10.5334/dsj-2017-002/.
- ↑ Lehnert, Kerstin; Wyborn, Lesley; Klump, Jens (2019). "FAIR Geoscientific Samples and Data Need International Collaboration" (in en). Acta Geologica Sinica - English Edition 93 (S3): 32–33. doi:10.1111/1755-6724.14236. ISSN 1755-6724. https://onlinelibrary.wiley.com/doi/abs/10.1111/1755-6724.14236.
- ↑ 26.0 26.1 Stall, Shelley; Yarmey, Lynn; Cutcher-Gershenfeld, Joel; Hanson, Brooks; Lehnert, Kerstin; Nosek, Brian; Parsons, Mark; Robinson, Erin et al. (1 June 2019). "Make scientific data FAIR" (in en). Nature 570 (7759): 27–29. doi:10.1038/d41586-019-01720-7. https://www.nature.com/articles/d41586-019-01720-7.
- ↑ Wilkinson, Mark D.; Dumontier, Michel; Aalbersberg, IJsbrand Jan; Appleton, Gabrielle; Axton, Myles; Baak, Arie; Blomberg, Niklas; Boiten, Jan-Willem et al. (15 March 2016). "The FAIR Guiding Principles for scientific data management and stewardship" (in en). Scientific Data 3 (1): 160018. doi:10.1038/sdata.2016.18. ISSN 2052-4463. PMC PMC4792175. PMID 26978244. https://www.nature.com/articles/sdata201618.
- ↑ 28.0 28.1 28.2 Guralnick, Robert P.; Cellinese, Nico; Deck, John; Pyle, Richard L.; Kunze, John; Penev, Lyubomir; Walls, Ramona; Hagedorn, Gregor et al. (4 June 2015). "Community Next Steps for Making Globally Unique Identifiers Work for Biocollections Data" (in en). ZooKeys 494: 133–154. doi:10.3897/zookeys.494.9352. ISSN 1313-2970. PMC PMC4400380. PMID 25901117. https://zookeys.pensoft.net/article/5042/.
- ↑ DataCite Metadata Working Group (2019). DataCite Metadata Schema for the Publication and Citation of Research Data v4.2. Madeleine de Smaele, Amy Hatfield Hart, Jan Ashton, Isabel Bernal Martinez, Stefanie Dietiker, Jannean Elliot. doi:10.5438/RV0G-AV03. http://schema.datacite.org/meta/kernel-4.2/.
- ↑ DCMI Usage Board (20 January 2020). "DCMI Metadata Terms". Dublin Core Metadata Initiative. DCMI. https://www.dublincore.org/specifications/dublin-core/dcmi-terms/. Retrieved 16 September 2020.
- ↑ 31.0 31.1 Cox, Simon Jonathan David (2011) (in en). ISO 19156:2011 - Geographic information -- Observations and measurements. International Organization for Standardization. doi:10.13140/2.1.1142.3042. http://rgdoi.net/10.13140/2.1.1142.3042.
- ↑ Group, Darwin Core Task (8 November 2014), "Darwin Core: 2014-11-08", Biodiversity Information Standards (TDWG) (Zenodo), doi:10.5281/zenodo.12694, https://zenodo.org/record/12694
- ↑ Reddy, T.B.K.; Thomas, Alex D.; Stamatis, Dimitri; Bertsch, Jon; Isbandi, Michelle; Jansson, Jakob; Mallajosyula, Jyothi; Pagani, Ioanna et al. (27 October 2014). "The Genomes OnLine Database (GOLD) v.5: a metadata management system based on a four level (meta)genome project classification". Nucleic Acids Research 43 (D1): D1099–D1106. doi:10.1093/nar/gku950. ISSN 1362-4962. PMC PMC4384021. PMID 25348402. https://doi.org/10.1093/nar/gku950.
- ↑ System For Earth Sample Registration (SESAR) (17 February 2020) (in en). SESAR XML Schema for samples. doi:10.5281/ZENODO.3875531. https://zenodo.org/record/3875531.
- ↑ IGSN (24 August 2017). "IGSN metadata". GitHub. https://github.com/IGSN/metadata.
- ↑ "Welcome to SESAR". SESAR. 2021. https://www.geosamples.org/.
- ↑ 37.0 37.1 Samy, Gaiji; Chavan, Vishwas; Ariño, Arturo H.; Otegui, Javier; Hobern, Donald; Sood, Rajesh; Robles, Estrella (9 July 2013). "Content assessment of the primary biodiversity data published through GBIF network: Status, challenges and potentials". Biodiversity Informatics 8 (2). doi:10.17161/bi.v8i2.4124. ISSN 1546-9735. http://dx.doi.org/10.17161/bi.v8i2.4124.
- ↑ 38.0 38.1 Robertson, Tim; Döring, Markus; Guralnick, Robert; Bloom, David; Wieczorek, John; Braak, Kyle; Otegui, Javier; Russell, Laura et al. (6 August 2014). "The GBIF Integrated Publishing Toolkit: Facilitating the Efficient Publishing of Biodiversity Data on the Internet" (in en). PLOS ONE 9 (8): e102623. doi:10.1371/journal.pone.0102623. ISSN 1932-6203. PMC PMC4123864. PMID 25099149. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0102623.
- ↑ 39.0 39.1 Ball-Damerow, Joan E.; Brenskelle, Laura; Barve, Narayani; Soltis, Pamela S.; Sierwald, Petra; Bieler, Rüdiger; LaFrance, Raphael; Ariño, Arturo H. et al. (11 September 2019). "Research applications of primary biodiversity databases in the digital age" (in en). PLOS ONE 14 (9): e0215794. doi:10.1371/journal.pone.0215794. ISSN 1932-6203. PMC PMC6738577. PMID 31509534. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0215794.
- ↑ "Global Biodiversity Information Facility". Global Biodiversity Information Facility. 2021. https://www.gbif.org/.
- ↑ Walls, Ramona L.; Deck, John; Guralnick, Robert; Baskauf, Steve; Beaman, Reed; Blum, Stanley; Bowers, Shawn; Buttigieg, Pier Luigi et al. (3 March 2014). "Semantics in Support of Biodiversity Knowledge Discovery: An Introduction to the Biological Collections Ontology and Related Ontologies" (in en). PLOS ONE 9 (3): e89606. doi:10.1371/journal.pone.0089606. ISSN 1932-6203. PMC PMC3940615. PMID 24595056. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0089606.
- ↑ 42.0 42.1 Buttigieg, Pier Luigi; Pafilis, Evangelos; Lewis, Suzanna E.; Schildhauer, Mark P.; Walls, Ramona L.; Mungall, Christopher J. (23 September 2016). "The environment ontology in 2016: bridging domains with increased scope, semantic density, and interoperation". Journal of Biomedical Semantics 7 (1): 57. doi:10.1186/s13326-016-0097-6. ISSN 2041-1480. PMC PMC5035502. PMID 27664130. https://doi.org/10.1186/s13326-016-0097-6.
- ↑ Osumi-Sutherland, D.; Zheng, J.; Buttigieg, P.L. et al. (n.d.). "Population and Community Ontology". https://raw.githubusercontent.com/PopulationAndCommunityOntology/pco/master/pco.owl.
- ↑ Avraham, Shulamit; Tung, Chih-Wei; Ilic, Katica; Jaiswal, Pankaj; Kellogg, Elizabeth A.; McCouch, Susan; Pujar, Anuradha; Reiser, Leonore et al. (1 January 2008). "The Plant Ontology Database: a community resource for plant structure and developmental stages controlled vocabulary and annotations". Nucleic Acids Research 36 (suppl_1): D449–D454. doi:10.1093/nar/gkm908. ISSN 0305-1048. PMC PMC2238838. PMID 18194960. https://academic.oup.com/nar/article/36/suppl_1/D449/2507667.
- ↑ 45.0 45.1 45.2 45.3 45.4 45.5 45.6 Damerow, Joan; Varadharajan, Charu; Boye, Kristin; Brodie, Eoin; Burrus, Madison; Chadwick, Dana; Cholia, Shreyas; Crystal-Ornelas, Robert et al.. (2020), "ESS-DIVE Global Sample Numbers and and Metadata Reporting Format for Environmental Systems Science (IGSN-ESS)" (in en), ESS-DIVE (Environmental System Science Data Infrastructure for a Virtual Ecosystem; Environmental Systems Science Data Infrastructure for a Virtual Ecosystem (ESS-DIVE)), doi:10.15485/1660470, https://www.osti.gov/servlets/purl/1660470/. Retrieved 2021-12-07
- ↑ ESS-DIVE (2021). "ESS-DIVE Sample ID and Metadata Reporting Format (IGSN-ESS) v1.1.0". GitHub. https://github.com/ess-dive-community/essdive-sample-id-metadata.
- ↑ 47.0 47.1 Toyoda, Jason G; Goldman, Amy E; Chu, Rosalie K; Danczak, Robert E; Daly, Rebecca A; Garayburu-Caruso, Vanessa A; Graham, Emily B; Lin, Xinming et al.. (2020), "WHONDRS Summer 2019 Sampling Campaign: Global River Corridor Surface Water FTICR-MS and Stable Isotopes" (in en), ESS-DIVE (Environmental System Science Data Infrastructure for a Virtual Ecosystem; Worldwide Hydrobiogeochemistry Observation Network for Dynamic River Systems (WHONDRS)), doi:10.15485/1603775, https://www.osti.gov/servlets/purl/1603775/. Retrieved 2021-12-07
- ↑ 48.0 48.1 Devaraju, Anusuriya; Klump, Jens; Tey, Victor; Fraser, Ryan; Cox, Simon; Wyborn, Lesley (2017), Kamps, Jaap; Tsakonas, Giannis; Manolopoulos, Yannis et al.., eds., "A Digital Repository for Physical Samples: Concepts, Solutions and Management", Research and Advanced Technology for Digital Libraries (Cham: Springer International Publishing) 10450: 74–85, doi:10.1007/978-3-319-67008-9_7, ISBN 978-3-319-67007-2, http://link.springer.com/10.1007/978-3-319-67008-9_7. Retrieved 2021-12-08
- ↑ Duerr, Ruth E.; Downs, Robert R.; Tilmes, Curt; Barkstrom, Bruce; Lenhardt, W. Christopher; Glassy, Joseph; Bermudez, Luis E.; Slaughter, Peter (1 September 2011). "On the utility of identification schemes for digital earth science data: an assessment and recommendations" (in en). Earth Science Informatics 4 (3): 139. doi:10.1007/s12145-011-0083-6. ISSN 1865-0473. https://link.springer.com/10.1007/s12145-011-0083-6.
- ↑ 50.0 50.1 50.2 Guralnick, Robert; Conlin, Tom; Deck, John; Stucky, Brian J.; Cellinese, Nico (3 December 2014). "The Trouble with Triplets in Biodiversity Informatics: A Data-Driven Case against Current Identifier Practices" (in en). PLOS ONE 9 (12): e114069. doi:10.1371/journal.pone.0114069. ISSN 1932-6203. PMC PMC4254916. PMID 25470125. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0114069.
- ↑ 51.0 51.1 51.2 51.3 51.4 51.5 51.6 51.7 Lehnert, Kerstin; Klump, Jens; Wyborn, Lesley; Ramdeen, Sarah (21 June 2019). "Persistent, Global, Unique: The three key requirements for a trusted identifier system for physical samples" (in en). Biodiversity Information Science and Standards 3: e37334. doi:10.3897/biss.3.37334. ISSN 2535-0897. https://biss.pensoft.net/article/37334/.
- ↑ 52.0 52.1 52.2 McMurry, Julie A.; Juty, Nick; Blomberg, Niklas; Burdett, Tony; Conlin, Tom; Conte, Nathalie; Courtot, Mélanie; Deck, John et al. (29 June 2017). "Identifiers for the 21st century: How to design, provision, and reuse persistent identifiers to maximize utility and impact of life science data" (in en). PLOS Biology 15 (6): e2001414. doi:10.1371/journal.pbio.2001414. ISSN 1545-7885. PMC PMC5490878. PMID 28662064. https://dx.plos.org/10.1371/journal.pbio.2001414.
- ↑ Michener, William K. (1 September 2015). "Ecological data sharing" (in en). Ecological Informatics 29: 33–44. doi:10.1016/j.ecoinf.2015.06.010. https://linkinghub.elsevier.com/retrieve/pii/S1574954115001004.
- ↑ Klump, Jens; Huber, Robert; Diepenbroek, Michael (1 March 2016). "DOI for geoscience data - how early practices shape present perceptions" (in en). Earth Science Informatics 9 (1): 123–136. doi:10.1007/s12145-015-0231-5. ISSN 1865-0473. http://link.springer.com/10.1007/s12145-015-0231-5.
- ↑ Ferguson, Christine; McEntrye, Jo; Bunakov, Vasily; Lambert, Simon; Sandt, Stephanie van der; Kotarski, Rachael; Stewart, Sarah; MacEwan, Andrew et al. (17 July 2018) (in en). D3.1 Survey Of Current Pid Services Landscape. doi:10.5281/ZENODO.1324296. https://zenodo.org/record/1324296.
- ↑ Goldstein, S.L.; Lehnert, K.A.; Hofmann, A.W. (29 January 2014). "IEDA Data DOI". doi.iedadata.org. http://doi.iedadata.org/100426. Retrieved 11 February 2019.
- ↑ 57.0 57.1 57.2 Walls, Ramona; Davies, Neil; Kansa, Sarah; Kunze, John; Lehnert, Kerstin; Vieglais, David (10 June 2020) (in en). Building transdisciplinary infrastructure for natural history material samples with the Internet of Samples (iSamples). doi:10.5281/ZENODO.4002440. https://zenodo.org/record/4002440.
- ↑ Lehnert, K. (17 December 2018). "IGSN: Toward a Mature and Generic Persistent Identifier for Samples". SlideShare. https://www.slideshare.net/klehnert/igsn-toward-a-mature-and-generic-persistent-identifier-for-samples. Retrieved 25 January 2019.
- ↑ McNutt, Marcia; Lehnert, Kerstin; Hanson, Brooks; Nosek, Brian A.; Ellison, Aaron M.; King, John Leslie (4 March 2016). "Liberating field science samples and data" (in EN). Science 351 (6277): 1024–26. doi:10.1126/science.aad7048. https://www.science.org/doi/abs/10.1126/science.aad7048.
- ↑ Kunze, J. (19 March 2021). "ARK Identifiers FAQ". ARKs in the Open Project. https://wiki.lyrasis.org/display/ARKs/ARK+Identifiers+FAQ.
- ↑ "Physical Sample Curation". ESIP Wiki. Federation of Earth Science Information Partners. 2021. https://wiki.esipfed.org/Physical_Sample_Curation.
- ↑ 62.0 62.1 62.2 Poisot, Timothée; Bruneau, Anne; Gonzalez, Andrew; Gravel, Dominique; Peres-Neto, Pedro (1 June 2019). "Ecological Data Should Not Be So Hard to Find and Reuse" (in English). Trends in Ecology & Evolution 34 (6): 494–496. doi:10.1016/j.tree.2019.04.005. ISSN 0169-5347. PMID 31056219. https://www.cell.com/trends/ecology-evolution/abstract/S0169-5347(19)30110-7.
- ↑ 63.0 63.1 Rauber, Andreas; Asmi, Ari; van Uytvanck, Dieter; Proell, Stefan (20 October 2015). Data Citation of Evolving Data: Recommendations of the Working Group on Data Citation (WGDC). doi:10.15497/rda00016. https://zenodo.org/record/1406002#.XG2FVOhKhaQ.
- ↑ Lehnert, K.A.; Klump, J.; Arko, R.A. et al. (December 2011). "IGSN e.V.: Registration and Identification Services for Physical Samples in the Digital Universe". American Geophysical Union, Fall Meeting 2011. Bibcode 2011AGUFMIN13B1324L. https://ui.adsabs.harvard.edu/abs/2011AGUFMIN13B1324L/abstract. Retrieved 01 March 2019.
- ↑ Michener, William K.; Brunt, James W.; Helly, John J.; Kirchner, Thomas B.; Stafford, Susan G. (1997). "Nongeospatial Metadata for the Ecological Sciences" (in en). Ecological Applications 7 (1): 330–342. doi:10.1890/1051-0761(1997)007[0330:NMFTES]2.0.CO;2. ISSN 1939-5582. https://onlinelibrary.wiley.com/doi/abs/10.1890/1051-0761%281997%29007%5B0330%3ANMFTES%5D2.0.CO%3B2.
- ↑ Voytek, Bradley (4 August 2016). Bourne, Philip E.. ed. "The Virtuous Cycle of a Data Ecosystem" (in en). PLOS Computational Biology 12 (8): e1005037. doi:10.1371/journal.pcbi.1005037. ISSN 1553-7358. PMC PMC4974004. PMID 27490108. https://dx.plos.org/10.1371/journal.pcbi.1005037.
- ↑ 67.0 67.1 67.2 67.3 67.4 Renaut, Sébastien; Budden, Amber E; Gravel, Dominique; Poisot, Timothée; Peres-Neto, Pedro (1 June 2018). "Management, Archiving, and Sharing for Biologists and the Role of Research Institutions in the Technology-Oriented Age" (in en). BioScience 68 (6): 400–411. doi:10.1093/biosci/biy038. ISSN 0006-3568. https://academic.oup.com/bioscience/article/68/6/400/4983937.
- ↑ Piwowar, Heather A.; Vision, Todd J. (1 October 2013). "Data reuse and the open data citation advantage" (in en). PeerJ 1: e175. doi:10.7717/peerj.175. ISSN 2167-8359. PMC PMC3792178. PMID 24109559. https://peerj.com/articles/175.
- ↑ Whitlock, Michael C. (1 February 2011). "Data archiving in ecology and evolution: best practices" (in en). Trends in Ecology & Evolution 26 (2): 61–65. doi:10.1016/j.tree.2010.11.006. https://linkinghub.elsevier.com/retrieve/pii/S0169534710002697.
- ↑ Peters, Debra P. C.; Loescher, Henry W.; SanClements, Michael D.; Havstad, Kris M. (1 March 2014). "Taking the pulse of a continent: expanding site-based research infrastructure for regional- to continental-scale ecology" (in en). Ecosphere 5 (3): art29. doi:10.1890/ES13-00295.1. ISSN 2150-8925. http://doi.wiley.com/10.1890/ES13-00295.1.
- ↑ Cox, Simon J. D. (1 January 2017). "Ontology for observations and sampling features, with alignments to existing models" (in en). Semantic Web 8 (3): 453–470. doi:10.3233/SW-160214. ISSN 1570-0844. https://content.iospress.com/articles/semantic-web/sw214.
- ↑ 72.0 72.1 Esteva, Maria; Walls, Ramona L.; Magill, Andrew B.; Xu, Weijia; Huang, Ruizhu; Carson, James; Song, Jawon (19 March 2019). "Identifier Services: Modeling and Implementing Distributed Data Management in Cyberinfrastructure" (in en). Data and Information Management 3 (1): 26–39. doi:10.2478/dim-2019-0002. ISSN 2543-9251. https://www.sciendo.com/article/10.2478/dim-2019-0002.
- ↑ Webb, Campbell O.; Ackerly, David D.; McPeek, Mark A.; Donoghue, Michael J. (1 November 2002). "Phylogenies and Community Ecology" (in en). Annual Review of Ecology and Systematics 33 (1): 475–505. doi:10.1146/annurev.ecolsys.33.010802.150448. ISSN 0066-4162. https://www.annualreviews.org/doi/10.1146/annurev.ecolsys.33.010802.150448.
- ↑ Jetz, W.; Thomas, G. H.; Joy, J. B.; Hartmann, K.; Mooers, A. O. (1 November 2012). "The global diversity of birds in space and time" (in en). Nature 491 (7424): 444–448. doi:10.1038/nature11631. ISSN 0028-0836. http://www.nature.com/articles/nature11631.
- ↑ Kelling, Steve; Hochachka, Wesley M.; Fink, Daniel; Riedewald, Mirek; Caruana, Rich; Ballard, Grant; Hooker, Giles (1 July 2009). "Data-intensive Science: A New Paradigm for Biodiversity Studies" (in en). BioScience 59 (7): 613–620. doi:10.1525/bio.2009.59.7.12. ISSN 0006-3568. https://academic.oup.com/bioscience/article-lookup/doi/10.1525/bio.2009.59.7.12.
- ↑ Rocca-Serra, P.; Sansone, S.-A.; Brandizi, M. (24 November 2008). "Specification documentation: Release candidate 1, ISA-TAB 1.0" (PDF). http://isatab.sourceforge.net/docs/ISA-TAB_release-candidate-1_v1.0_24nov08.pdf.
- ↑ Horsburgh, Jeffery S.; Aufdenkampe, Anthony K.; Mayorga, Emilio; Lehnert, Kerstin A.; Hsu, Leslie; Song, Lulin; Jones, Amber Spackman; Damiano, Sara G. et al. (1 May 2016). "Observations Data Model 2: A community information model for spatially discrete Earth observations" (in en). Environmental Modelling & Software 79: 55–74. doi:10.1016/j.envsoft.2016.01.010. https://linkinghub.elsevier.com/retrieve/pii/S1364815216300093.
- ↑ Diepenbroek, Michael; Grobe, Hannes; Reinke, Manfred; Schindler, Uwe; Schlitzer, Reiner; Sieger, Rainer; Wefer, Gerold (1 December 2002). "PANGAEA—an information system for environmental sciences" (in en). Computers & Geosciences 28 (10): 1201–1210. doi:10.1016/S0098-3004(02)00039-0. https://linkinghub.elsevier.com/retrieve/pii/S0098300402000390.
- ↑ Gardner, Timothy (22 August 2014). "A swan in the making" (in en). Science 345 (6199): 855–855. doi:10.1126/science.1259740. ISSN 0036-8075. https://www.science.org/doi/10.1126/science.1259740.
- ↑ Teal, Tracy K.; Cranston, Karen A.; Lapp, Hilmar; White, Ethan; Wilson, Greg; Ram, Karthik; Pawlik, Aleksandra (18 March 2015). "Data Carpentry: Workshops to Increase Data Literacy for Researchers". International Journal of Digital Curation 10 (1): 135–143. doi:10.2218/ijdc.v10i1.351. ISSN 1746-8256. http://www.ijdc.net/article/view/10.1.135.
- ↑ Enke, Neela; Thessen, Anne; Bach, Kerstin; Bendix, Jörg; Seeger, Bernhard; Gemeinholzer, Birgit (1 September 2012). "The user's view on biodiversity data sharing — Investigating facts of acceptance and requirements to realize a sustainable use of research data —" (in en). Ecological Informatics 11: 25–33. doi:10.1016/j.ecoinf.2012.03.004. https://linkinghub.elsevier.com/retrieve/pii/S1574954112000222.
- ↑ 82.0 82.1 82.2 Freedman, Leonard P.; Cockburn, Iain M.; Simcoe, Timothy S. (9 June 2015). "The Economics of Reproducibility in Preclinical Research" (in en). PLOS Biology 13 (6): e1002165. doi:10.1371/journal.pbio.1002165. ISSN 1545-7885. PMC PMC4461318. PMID 26057340. https://dx.plos.org/10.1371/journal.pbio.1002165.
- ↑ Page, Roderic (7 April 2016). "Towards a biodiversity knowledge graph". Research Ideas and Outcomes 2: e8767. doi:10.3897/rio.2.e8767. ISSN 2367-7163. http://rio.pensoft.net/articles.php?id=8767.
- ↑ Farrell, Joseph; Simcoe, Timothy (6 August 2012). Four Paths to Compatibility. Oxford University Press. doi:10.1093/oxfordhb/9780195397840.013.0002. http://oxfordhandbooks.com/view/10.1093/oxfordhb/9780195397840.001.0001/oxfordhb-9780195397840-e-2.
- ↑ Cousijn, Helena; Kenall, Amye; Ganley, Emma; Harrison, Melissa; Kernohan, David; Lemberger, Thomas; Murphy, Fiona; Polischuk, Patrick et al. (1 December 2018). "A data citation roadmap for scientific publishers" (in en). Scientific Data 5 (1): 180259. doi:10.1038/sdata.2018.259. ISSN 2052-4463. PMC PMC6244190. PMID 30457573. http://www.nature.com/articles/sdata2018259.
- ↑ Hanson, Brooks (7 January 2016). "AGU Opens Its Journals to Author Identifiers". Eos 97. doi:10.1029/2016EO043183. ISSN 2324-9250. https://eos.org/agu-news/agu-opens-its-journals-to-author-identifiers.
- ↑ Lin, Jennifer; Strasser, Carly (28 October 2014). "Recommendations for the Role of Publishers in Access to Data" (in en). PLoS Biology 12 (10): e1001975. doi:10.1371/journal.pbio.1001975. ISSN 1545-7885. PMC PMC4211645. PMID 25350642. https://dx.plos.org/10.1371/journal.pbio.1001975.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. The original article lists references in alphabetical order; however, this version lists them in order of appearance, by design.