User:Shawndouglas/sandbox/sublevel12
|
|
This is sublevel12 of my sandbox, where I play with features and test MediaWiki code. If you wish to leave a comment for me, please see my discussion page instead. |
Sandbox begins below
Title: Laboratory Informatics: Information and Workflows
Author for citation: Joe Liscouski
License for content: Creative Commons Attribution-ShareAlike 4.0 International
Publication date: April 2024
NOTE: This content originally appeared in Liscouski's Computerized Systems in the Modern Laboratory: A Practical Guide as Chapter 3 - Laboratory Informatics / Departmental Systems, published in 2015 by PDA/DHI, ISBN 193372286X. This is reproduced here with the author's / copyright holder's permission. Some changes have been made to the original material, replacing some out-of-date screen shots with vendor-neutral mock-ups, for example. In addition, note that some specifications for network speeds are out-of-date but the concerns with system performance are still realistic.
Introduction
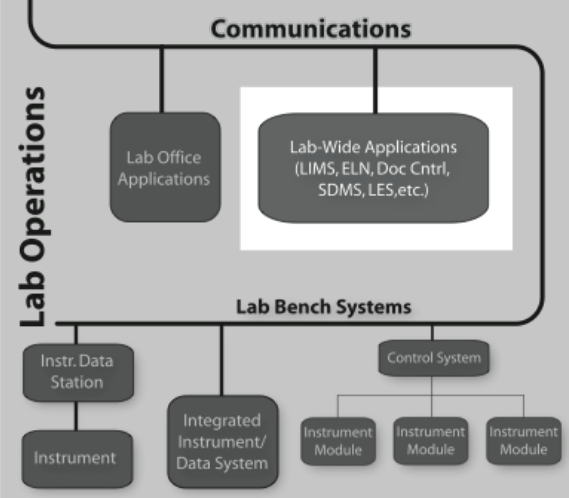
Laboratory informatics refers to software systems that usually are accessible at the departmental level–they are often shared between users in the lab-and focus on managing lab operations, and lab-wide information rather than instrument management or sample preparation (Figure 3-1).
|
Differing from standard office applications (e.g., word processing, spreadsheets, etc.), laboratory informatics software solutions include the:
- laboratory information management system (LIMS) and laboratory information system (LIS)
- electronic laboratory notebook (ELN)
- document management system (DMS)
- scientific data management system (SDMS)
- laboratory execution system (LES), and
- chemical inventory management (CIM).
Before we get into the details of what these technologies are, we must establish a framework for understanding laboratory operations so that we can see where products fit in the lab's workflow. The products you introduce into your lab are going to depend on the lab's needs, and given the complexity of vendor offerings and their potential interactions and overlapping capabilities, defining your requirements is going to take some thought.
These products are undergoing a rapid evolution driven by market pressures as vendors compete for your business, partly by trying to cover as much of a lab’s operations as they can. If you look at functional checkboxes in brochures, they often cover similar elements, but their strengths, weaknesses, and methods of operation are different, and those differences should be important to you.
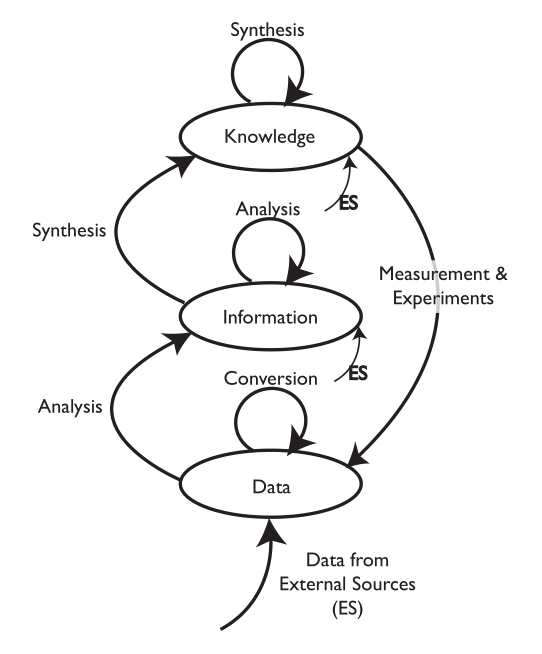
We are going to start by comparing two different types of laboratory environments: research labs and service labs (laboratories whose role is to provide testing, and assays, for example, quality control, clinical labs, etc.). That comparison and subsequent material is going to be done with the aid of an initially simple model for the development and flow of knowledge, information, and data within laboratories (Figure 3-2).
|
The model consists of ovals and arrows; the ovals represent collections of files and databases supporting applications, and the arrows are processes for working with the materials in those collections. The “ES” abbreviation notes processes that bring in elements from sources outside the lab. The first point we need to address is the definition of “knowledge,” “information,” and “data,” as used here. We are not talking about philosophical points, but how these are represented and used in the digital world. “Knowledge” is usually reports, documents, etc. that may exist as individual files or be organized and accessed through applications software. That software would include DMSs, databases for working with hazardous materials, reference databases, and access to published material stored locally or accessed over internal/external networks.
“Information” consists of elements that can be understood by themselves such as pH measurements, the results of an analysis, an object's temperature, an infrared spectrum, the name of a file, and so on. Information elements can reside as fields in databases and files. Information can be provided as meaningful answers to questions. Finally, “Data” refers to measurements that by themselves may not have any meaning, or that require conversion or be combined with other data and analyzed before they are useful information. For example, the twenty-seventh value in a digital chromatogram data stream, the area of a peak (needs comparison to known quantities before it is useful), millivolt readings from a pH meter (you need temperature information and other elements before you can convert it to pH), etc.
There are grey areas and examples where these definitions may not hold up well, but the key point is how they are obtained, stored, and used. You may want to modify the definitions to suit your work, but the comments above are how they are used here.
The arrows represent processes that operate on material in one storage structure and put the results of the work in another. Processes will consist of one or more steps or stages and can include work done in the lab as well as outside the lab (outsourced processing, access to networked systems, for example). It is possible that operations will be performed on material in one storage structure and have those results placed in the same storage level. For example, several “information” elements may be processed to create a new information entity.
Initially, we are going to discuss the model as a two-dimensional structure, but those storage systems and processes have layers of software, hardware, and networked communications. In addition, the diagram as shown is a simplification of reality since it shows only one process for an experiment. In the real world, there would be a process line for each laboratory process in your lab, and the “data” oval would represent a collection of data storage elements from each of the data acquisition/storage/analysis systems in the lab. Each experimental process would have a link to the “knowledge” structures catalog of standard operating procedures (SOPs). As we start to build on this structure we can see how requirements for products and workflow can be derived.
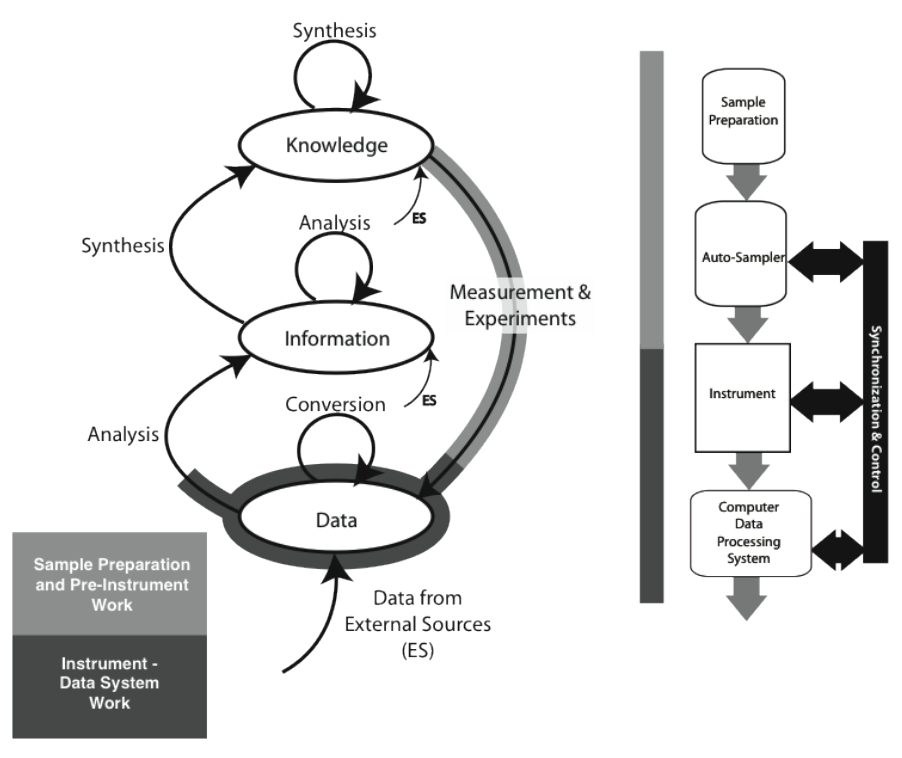
The material covered in the Lab Bench chapter (Chapter 1) fits the model as shown in Figure 3-3 (the model used in the Lab Bench discussion is shown as well; it is represented by the light-/heavy-grey lines in the K/I/D model). The sample preparation and pre-instrumental analysis work is shown as the light-grey / heavy-line portion of the “Measurement & Experiments” process, with instrumental data acquisition, storage, and analysis displayed with the darker grey/heavy line (which may be entirely or partly completed by the software; the illustration shows the “partial” case). Not all experiment/instrument interactions result in the use of a storage system. Balances, for example, may hold one reading and then send it through to the information storage on command, where it may be analyzed with other information. The procedure description needed to execute a laboratory process—the measurement and experiment—would come from material in the “Knowledge” storage structure.
|
What we are concerned about in laboratory informatics is what happens to the data and information that results either from data analysis or information gained directly from experiments.
Comparing two different laboratory operational environments
By its nature, research work can be varied. SOPs may change weekly or more slowly, and the information collected can change from one experiment to another. The information collected can be in the form of tables, discrete reading, images, text, video, audio, or other types of information. The work may change depending on the course of the research project. Aside from product support on the lab bench, the primary need is for a means of recording the progress of projects and managing the documents connected with the work.
Until recently, that need was met through the use of paper notebooks, with entries dated, signed by the author, and counter-signed by a witness. The researcher would document the work by handwritten entries, and instrument output would be recorded similarly or have printouts taped to notebook pages. Anything that could not be printed and pasted in would have references entered. There are a few problems with this approach:
- Handwritten records can be difficult to decipher.
- Paper is subject to deterioration by a variety of methods.
- Taped/pasted entries can come loose and be lost.
- References to files, tapes, instrument recordings, etc. that are stored separately can become difficult to track if the material is moved.
- “Backup” can be a problem since you are working with physical media; the obvious step is to make copies of every page after it has been signed and witnessed.
- The notebook contents, particularly if the notebook has been archived, are only useful for as long as someone remembers that they exist and can provide information that helps in locating the entries. Most labs have stories that start “I remember someone doing that work, but…” and the matter is either dropped or the work repeated.
The intellectual property recorded in notebooks is of value only if someone knows it exists, and that it can be found and understood. There is another point that needs to be mentioned here that we will reference later: lab personnel consider entries in paper lab notebooks as “their work and their information.” To the extent that it represents their efforts, they are right, but when it comes to access control and ownership, the contents belong to whoever paid for the work.
Research work is sometimes done by a single individual, but often it has several people working together in the same lab or collaborating with researchers in other facilities. Those cooperative programs may be based on:
- Researchers working on independent projects from a common database of information on biological materials, chemical compounds and their effect on bacteria or viruses, toxicology, pharmacology, pharmacokinetics, reaction mechanisms, etc. in life sciences, data gathered from particle collisions in physics, chemical structures in chemistry, and so on.
- Researchers working in collaborative programs where the outcomes are co- authored reports, presentations, etc.
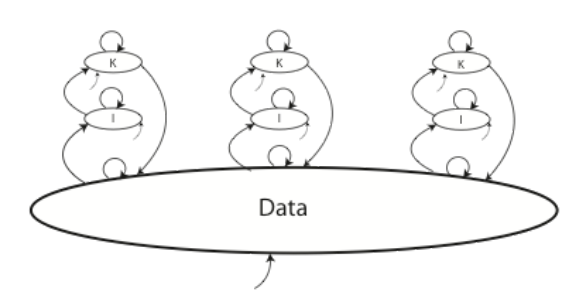
Whether we are looking at single individuals or cooperative work, each of those situations has an impact on the way knowledge, information, and data is collected, developed, shared, and used. For example, take a look at Figure 3-4 below.
|
The figure shows three researchers working on individual projects, contributing to, and working from, a common data set; this could in fact be one data system or a collection of data systems from different sources. The bottom arrow represents data coming in from some outside source.
Regardless of whether this is an electronic system or a paper-based process, there still is the following set of requirements:
- There has to be a catalog of what records are in the system, as people need the ability to read the catalog, search it, and add new material. Removing material from the catalog would be prohibited for two reasons. First, someone might be using the material and deletion would create problems. Second, if you are working in a regulated environment, deleting material is not permitted. Making this work means that someone is going to be tasked as the system administrator.
- Material cannot be edited or modified. If changes are needed, the original material remains as-is, and a new record with the changed material is created that would contain a description of the changes, why they were made, who is responsible, and when the work was done (i.e., via an audit trail). This creates a parent-child relationship between data elements that could become a lengthy chain. One simple example can be found with imaging. An image of something is taken and stored, and any enhancements or modifications would create a set of new records linked back to the original material. This is an audit trail with all the requirements that are carried with it.
- The format for files containing material should be standardized. Having similar types of material in differing file structures can significantly increase the effort and ability to work with the data (data in this case, with the same issues extending to information and knowledge). If you have two or more people working with similar instruments, having the data stored in the same format is preferable to having different formats from different vendors. Plate readers (for microplates) usually use CSV file formats; however, instruments such as chromatographs, spectrometers, etc. will use different file formats depending on the vendors. If this is the case, you may have to export material in a neutral format for shared access. At this point in time, the user community has not developed the standardized file format for instrumentation to make this possible, hence the use of “should” earlier in this bullet rather than a stronger statement. That could change with the finalization of the ANiML standard (ASTM WK23265) for analytical data. Users can standardize the format within their organization by working within a vendor’s product family.
- The infrastructure for data backups should be instituted to protect the material and access to it. This point alone would shift the implementation toward electronic data management because of the ease with which this can be done. Data collection represents a significant investment in resources, with a corresponding value, and backup copies (local and remote) are just part of good planning.
- Policies have to be defined about when and how archiving takes place. Material may be old, but still referenced, and removing it from the system will create issues. If the implementation is electronic, storage systems are becoming inexpensive, so expanding storage should not be a problem.
- The system must have sufficient security mechanisms to protect from unauthorized access and electronic intrusion.
About the author
Initially educated as a chemist, author Joe Liscouski (joe dot liscouski at gmail dot com) is an experienced laboratory automation/computing professional with over forty years of experience in the field, including the design and development of automation systems (both custom and commercial systems), LIMS, robotics and data interchange standards. He also consults on the use of computing in laboratory work. He has held symposia on validation and presented technical material and short courses on laboratory automation and computing in the U.S., Europe, and Japan. He has worked/consulted in pharmaceutical, biotech, polymer, medical, and government laboratories. His current work centers on working with companies to establish planning programs for lab systems, developing effective support groups, and helping people with the application of automation and information technologies in research and quality control environments.
References