Journal:Conversion of a classical microbiology laboratory to a total automation laboratory enhanced by the application of lean principles
| Full article title | Conversion of a classical microbiology laboratory to a total automation laboratory enhanced by the application of lean principles |
|---|---|
| Journal | Microbiology Spectrum |
| Author(s) | Trigueiro, Graça; Oliveira, Carlos; Rodrigues, Alexandra; Seabra, Sofia; Pinto, Rui; Bala, Yohann; Granado, Monica G.; Vallejo, Sandra; Gonzalez, Victoria; Cardoso, Carlos |
| Author affiliation(s) | Dr. Joaquim Chaves Clinical Analysis Laboratory, bioMérieux |
| Primary contact | Email: alexandra dot cristina at jcs dot pt |
| Editors | Doucet-Populaire, Florence C. |
| Year published | 2024 |
| Volume and issue | 12(2) |
| Article # | e02153-23 |
| DOI | 10.1128/spectrum.02153-23 |
| ISSN | 2165-0497 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://journals.asm.org/doi/10.1128/spectrum.02153-23 |
| Download | https://journals.asm.org/doi/pdf/10.1128/spectrum.02153-23?download=true (PDF) |
Abstract
Laboratory automation in microbiology improves productivity and reduces sample turnaround times (TATs). However, its full potential can be unlocked through the optimization of workflows by adopting lean principles. This study aimed to explore the relative impact of laboratory automation and continuous improvement events (CIEs) on productivity and TATs. Laboratory automation took place in November 2020 and consisted of the introduction of WASPLab and VITEK MS systems. CIEs were run in May and September 2021. Before the conversion, the laboratory processed approximately 492 samples on weekdays and had 10 full-time equivalent (FTE) staff for a productivity of 49 samples/FTE/day. In March 2021, after laboratory automation, the caseload went up to approximately 621 while the FTEs decreased to 8.5, accounting for a productivity improvement to 73 samples/FTE/day. The hypothetical productivity went up to 110 samples/FTE/day following CIEs, meaning that the laboratory could at that point deal with a caseload increase to approximately 935 with unchanged FTEs. Laboratory conversion also led to an improvement in TATs for all sample types. For vaginal swabs and urine samples, median TATs decreased from 70.3 hours (interquartile range [IQR]: 63.5–93.1) and 73.7 hours (IQR: 35.6–50.7) to 48.2 hours (IQR: 44.8–67.7) and 40.0 hours (IQR: 35.6–50.7), respectively. Automation alone was responsible for 37.2% and 75.8% of TAT reduction, respectively, while the remaining reduction of 62.8% and 24.2%, respectively, was achieved due to CIEs. The laboratory reached productivity and TAT goals predefined by the management after CIEs. In conclusion, automation substantially improved productivity and TATs, while the subsequent implementation of lean management further unlocked the potential of laboratory automation.
Importance
In this study, we combined total laboratory automation with lean management to show that appropriate laboratory work organization enhanced the benefit of the automation and substantially contributed to productivity improvements. Globally, the rapid availability of accurate results in the setting of a clinical microbiology laboratory is part of patient-centered approaches to treat infections and helps the implementation of antibiotic stewardship programs backed by the World Health Organization (WHO). Locally, from the point of view of laboratory management, it is important to find ways of maximizing the benefits of the use of technology, as total laboratory automation is an expensive investment.
Keywords: turnaround time, productivity, lean principles, laboratory automation, change management
Introduction
Compared to general hematology and biochemistry laboratories, automation was late to arrive in microbiology laboratories because of the complexity of the processes involved.[1][2][3] The most important driver of automation adoption in microbiology laboratories in recent times is represented by a shift of paradigm in clinical microbiology that increased the demand for rapid and accurate results.[1][2][4] The availability of such results is important for the success of patient-centered approaches to treat infections and for antibiotic stewardship programs.[5] Over the past 20 years, and especially since the onset of the coronavirus disease 2019 (COVID-19) pandemic, microbiology laboratories have been increasingly adopting technologies and processes that improve quality and efficiency, alongside a reduction in sample turnaround times (TATs).[1]
In total laboratory automation systems, all main system components (i.e., plating/streaking unit, incubation unit, high-resolution imaging system, colony picking for identification and antimicrobial susceptibility testing, and post-imaging analysis workstation) are robot driven. Artificial intelligence (AI) has recently been incorporated into imaging analysis to automatically interpret plates. Automating analyses has been the key to improving productivity.[6][7][8] System components may be connected via a conveyor belt system or placed in convenient locations in laboratories.[9] In the WASPLab system, plates are sorted out to stackers and transferred to workstations by technologists. The systems must be flexible enough to accommodate several variables, such as specimen types and non-standardized containers. Moreover, some samples are sent to be screened for a specific pathogen (e.g., throat swab samples to be screened for Streptococcus pyogenes), while others may require a variety of microbial culture and identification methods for a full diagnostic work-up.[1]
There are numerous advantages to total laboratory automation. Although a high investment is needed for the conversion from a manual to an automated lab, in the long run, automated laboratories are cheaper to run.[4] The implementation of total laboratory automation increases efficiency, measured as turnaround time and total productivity, and improves the quality of testing, thanks to the standardization and optimization of the procedures. Automation offers a possibility of better sample management and traceability, often requires lower volumes of samples, renders laboratory accreditation easier to obtain, and lowers the biological risk to the operators.[2][10] In the context of a microbiology laboratory, a significantly better recovery of pathogens—defined as a greater number of unique colony morphologies, a higher proportion of discrete colonies, and the identification of more species/plates—can be achieved, as described by Bailey et al.[1]
However, there are two principal elements of laboratory automation: hardware and workflow[11]. Total laboratory automation improves efficiency through the reduction of repetitive tasks with moderate added value, i.e., through the modification of the workflow. Automated incubators with digital imaging drastically reduce the number of manipulations of the culture media plates. About 97% of samples are suitable for automatic processing.[2] Employees often resist change. Fear of the unknown, lack of trust toward leaders, comfort derived from a familiar routine, lack of perception that a change is needed, perceived lack of knowledge or competence and the necessity to retrain, poor communication of what will happen from the management, and exhaustion or saturation when changes are too frequent are all factors at the basis of resistance to change.[12] Therefore, to be successful, the introduction of a change must be “managed” through a process known as change management. Change management occurs through steps (i.e., prepare, implement, monitor, sustain, and reevaluate).[12] In particular, a continuous improvement in streamlining workflow must be introduced. Change management is done with the use of lean methodology, an evidence-based approach to increase quality and efficiency that combines philosophy, processes, people, and structures.[13]
The study aimed to assess the relative impact of laboratory automation, change management, and continuous improvement events (CIEs) on key performance indicators (KPIs, i.e., full-time equivalent [FTE]/day and TAT) in a clinical microbiology laboratory concerning the processing of urine samples, vaginal swabs, and stool samples that comprise over 90% of the caseload of the laboratory.
Results
Pre-conversion
Caseload and FTE
In May 2019, prior to the conversion, Dr. Joaquim Chaves Saúde Clinical Analysis Laboratory (JCS) processed on average 492 samples/day on weekdays, 389 on Saturdays, and approximately 12 on Sundays. Three-quarters of the samples received were urine samples, 16% were vaginal swabs, 4% stool samples, 2% pharyngeal swabs, and about 1% were samples sent in for mycology testing. Other sample types comprised 3%. The laboratory had 10 FTEs of staff at the time.
KPIs
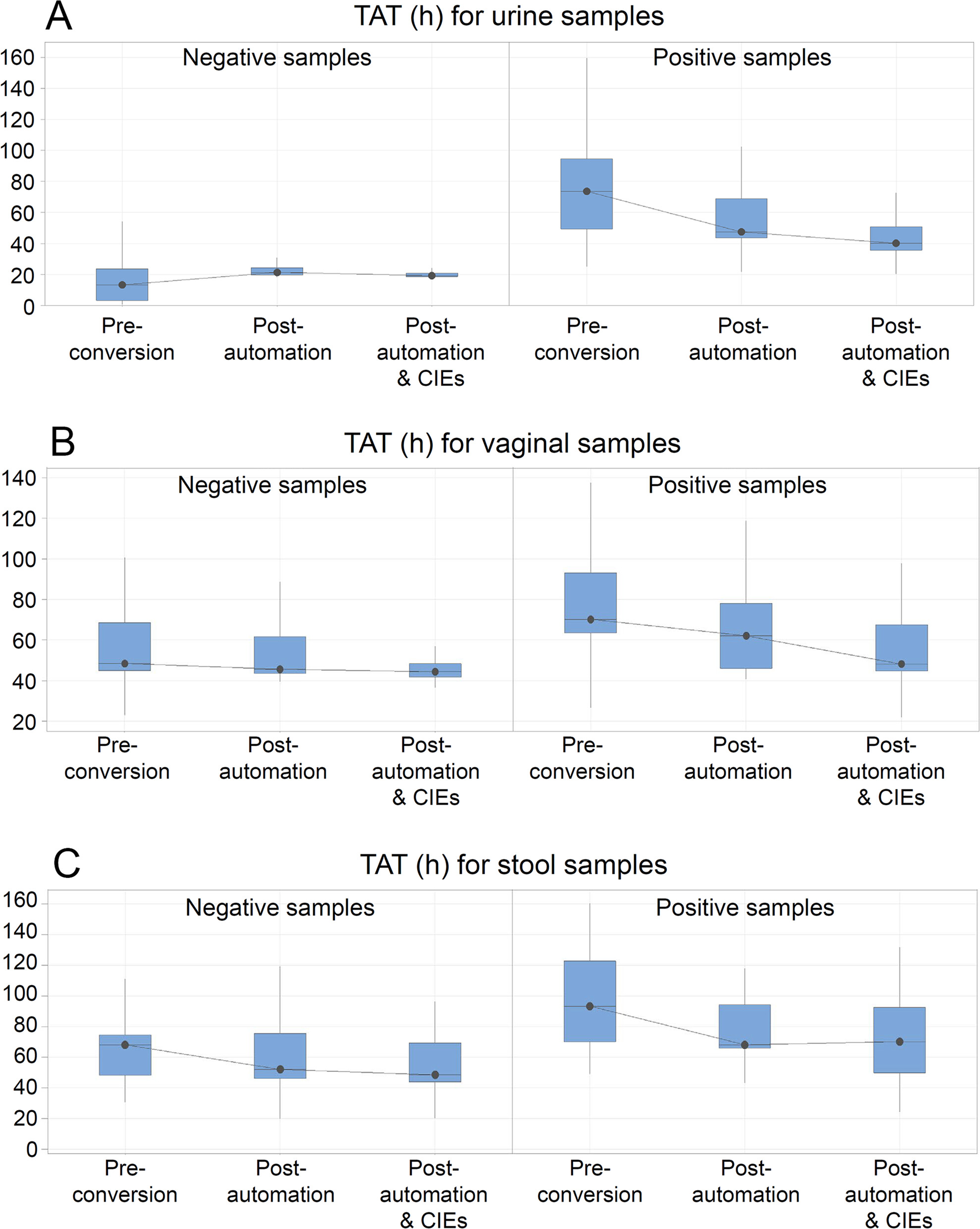
The productivity pre-conversion was 49 samples/FTE/day (Table 1). The median TAT was the shortest for the negative urine samples (median 13 hours; interquartile range IQR; 3.1; 23.7]) and the longest for the positive stool samples (median 93.3 hours; IQR [70.1; 122.8]). Positive urine and vaginal swabs, in comparison, had a similar median TAT with a narrower IQR for vaginal swabs (median 73.7 hours; IQR [43.4; 94.6] versus median 70.3 hours; IQR [64.5; 93.1]). Full results are given in Table 2.
| ||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Post-automation
Caseload and FTE
In March 2021, following the introduction of automation, the average number of samples in a day processed by JCS equaled 621 on weekdays, 417 on Saturdays, and about11 on Sundays. The distribution of sample types changed only slightly. Seventy-eight percent of samples received were urine samples, 12% were vaginal swabs, 2% were stools, and 1% were pharyngeal swabs, and approximately 1% were samples sent in for mycology testing. Other sample types comprised 6%. In March 2021, the availability of FTEs in the laboratory went down to 8.5 (Table 1).
KPIs
Given the average number of samples received in a day and the reduced FTEs, the productivity of the JCS laboratory increased to 73 samples/FTE/day (Table 1). The automation led to a decrease in TAT for all sample types except for negative urine samples due to the discontinuation of the UF1000i urinalysis system, whereas the decrease by 26% of the TAT for positive stool samples did not reach statistical significance (P = 0.121). Nonetheless, the median TAT was still the shortest for the negative urine samples (median 21.3 hours; IQR [21.2; 22.9]) and the longest for the positive stool samples (median 69.5 hours; IQR [67.3; 94.4]). It is important to mention that for the urine workflow, the removal of the screening step using the UF1000 explains the longer TAT for negative urine, since currently, the sample status depends on the interpretation of culture on agar. TAT for positive urine samples went down to a median of 47.4 hours (43.5; 68.7), while positive vaginal swabs decreased to a median of 62.2 hours (46.1; 85.1) (Table 2).
Post-automation and CIEs
Caseload and FTE
In September 2021, the average number of samples processed and the available FTEs remained unchanged since March 2021.
KPIs
Automation, together with continuous improvement events, increased productivity to a theoretical level of 110/FTE/day, equivalent to processing 935 samples/day with the same FTEs, which more than doubled the original productivity observed pre-conversion (Table 1). Further improvement of median TAT was observed for positive urine (median 40.0 hours; IQR [35.6; 50.7]) and vaginal swab (median 48.2 hours; IQR [44.8; 67.7]) samples, while no additional improvement was seen for the positive stool samples. Predefined goals were reached for all samples except for positive stool specimens (Table 2).
The introduction of WASPLab and VITEK MS together with the CIEs led to a reduction in variability of TAT, shown as a decrease in the IQR range (Table 2). For the negative samples, an IQR reduction of 89%, 72%, and 3% was observed for urine, vaginal swab, and stool specimens, respectively, while the values equaled 67%, 23%, and 19% for the positive counterparts.
The relative impact of automation and CIEs on overall reduction in TAT
Laboratory conversion (automation and CIEs) led to a 124% improvement in productivity and improved TATs by a value ranging from 8% (for negative vaginal swabs) to 46% (for positive urine samples). TAT for positive vaginal swabs and stool samples improved by 31% and 26%, respectively (Table 2).
Discussion
The conversion of the JCS laboratory, consisting of laboratory automation facilitated by change management, substantially improved the productivity and TATs of the laboratory and reached goals predefined by the management. The improvement was the greatest for the positive urine samples, for which the median TAT decreased from just over three days (73.7 hours) to well under two days (40 hours, Fig. 1). The discontinuation of the UF1000 automated urine particle analyzer caused the expected lengthening of TAT for negative urine samples, although the post-conversion median TAT met the expected goal predefined by the JCS management. Moreover, TAT variability decreased following laboratory automation and CIEs. This occurred through an improvement of the standardization of the processes and full alignment with the pre-established priorities.
|
A more streamlined automatic workflow that eliminated many manual tasks provided the major contribution to TAT reduction; however, CIEs maximized the potential benefit of laboratory automation and contributed substantially to productivity improvements. In 2019, the lab was processing, on average, 492 samples a day with 10 FTEs; after laboratory automation, the lab was processing 621 samples a day with 8.5 FTEs. After CIEs, with 8.5 FTEs, the lab could process 935 samples per day. In 2019, 19 FTEs would have been needed to process a daily caseload of 935 specimens, whereas after the integration of laboratory automation, 13 FTEs would have been needed. In other words, had there been a caseload of 935 at the time, the improvement in productivity observed following lab conversion and optimization of workflows would have allowed for the saving of six FTEs attributable to laboratory automation alone, and of an additional 4.5 FTEs made possible by the application of lean principles (i.e., the CIEs).
The delivery of the change management program in JCS helped the laboratory staff to increase their level of understanding of automation. The change management program focused on defusing reluctance to technological changes, motivated the adoption of WASP and WASPLab technologies, developed confidence toward hardware and software systems, envisioned daily work with automation, worked on the understanding that skills are transformed and not lost, and clarified the expectations and decisions to be made within the laboratory. The procedure optimization phase based on lean management application allowed the team of the Department of Microbiology at the JCS to develop a mindset of continuous improvement that is needed to maximize the potential of laboratory automation, reach preset goals, and to sustain them, which is a fundamental element of lean management.[14]
The results of this study confirm the findings published by others. In the setting of a microbiology laboratory, laboratory automation has historically been shown to improve efficiency.[15][16][17][18][19][20][21][22] In addition, the combination of matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF)-based microbial identification and total laboratory automation individually and together improved TATs in microbiology.[23][24] Da Rin and colleagues showed that the integration of the WASPLab diagnostic microbiology system into a pre-existing system of total automation, together with resource standardization and optimization, shortened TAT.[25] In a Chinese study, total laboratory automation was accompanied by the setting up of three working shifts, as opposed to a single working shift pre-automation. Together, the two actions led to a statistically significant shortening of TATs, especially for the cerebrospinal fluid sample diagnostics.[26]
Unlike Cherkaoui and colleagues, who saw only a trend toward shorter median TATs, our intervention led to a substantial improvement in TAT for positive urine samples. This result strongly supports the notion that automation must be matched by laboratory work organization, which was a limitation acknowledged by Cherkaoui et al. in their study.[27] In our study, laboratory automation alone was responsible for 37.2% and 75.8% reductions in TAT observed for positive vaginal swabs and positive urine samples, respectively, while the remaining 62.8% and 24.2% reduction in TAT for these samples was achieved due to CIEs. The study by Yarborough et al. confirmed that changes in urine sample workflow were necessary to maximize the efficiency of laboratory automation and to optimize TAT.[28]
Taken together, the conversion of the JCS laboratory resulted in several benefits. The first benefit was the occurrence of fewer errors and associated rework. Most of the laboratory errors occur during the extra (pre- and post-) analytical phases of the testing process.[29] Indeed, in the workflow before the conversion, there was a lot of room for errors since some of the samples were labeled manually and test results were manually fed into the laboratory information system (LIS). Secondly, the conversion led to fewer uncertainties concerning operating procedures, which in turn counteracted the feeling of being overwhelmed, a sensation that may be experienced by laboratory staff after a major change to the working routine. Thirdly, during the conversion, an environment favorable to continuous improvement was created, with clear procedures allowing any problem or deviation to be easily spotted. In addition, sample type-based CIEs enabled focused collaboration of the technologists and sequential validation. Such sample type-based optimization was also done by Cherkaoui et al. in Geneva.[30] Lastly, productivity improvements facilitated qualified staff transfer to clinical activities despite working in the context of the COVID-19 pandemic, which led to a 20% caseload increase.
At the level of global trends, the introduction of laboratory automation and CIE helps counteract problems with staffing experienced by most of the clinical laboratories worldwide due to accelerated rates of microbiologists’ retirement and fewer people entering the field of laboratory medicine. A very recent study showed that up to 80% of microbiology laboratories have vacancies and struggle to fill them.[31] There is also the phenomenon of declining reimbursement, and total laboratory automation is a way of obtaining cost savings.[32]
There is one main limitation to the study, which is that it is the experience of one center. While laboratory automation has been demonstrated to provide benefits regardless of site size/organization[4], the magnitude of the benefit and impact of CIEs are likely to be site-dependent since they require finding the right combination of people, processes (i.e., workflow), and technology to unlock new sources of improvement.[11] Moreover, the productivity index used in the study is strongly linked to the relative local distribution of sample types, which at the JCS laboratory was 74% urine samples, 16% vaginal swabs, 4% stool samples, and 6% other specimens. Consequently, the improvements in productivity observed after lab automation and after CIEs would be expected to vary with a different sample distribution.
A minor limitation of the study is the fact that the workflow for positive stool samples did not reach the predefined TAT goal of 65 hours. However, stool samples represent only 4% of the caseload, and only 5% of them are positive, which makes the standardization of the workflow for these isolated specimens difficult. Furthermore, while previous studies by Cherkaoui and colleagues also highlighted the potential of automation to reduce TAT for other critical samples such as blood culture[33], the caseload for this sample type in the JCS laboratory was too low to be included in the present study. Moreover, the present study looked at the impact of a bundled intervention, i.e., implementation of WASPLab and VITEK MS systems. While the decrease of TAT is most likely caused by the combination of both technologies, the relative impact of each of the changes was not assessed.
Conclusion
In conclusion, JCS conversion resulted in substantial improvements in KPIs. While automation alone substantially improved TAT and productivity, the subsequent implementation of lean management further unlocked the potential of laboratory automation through the streamlining of the processes involved. Together, they led to the achievement of predefined goals. Future studies will be needed to see how additional developments in the field of automation—e.g., automatic colony picking for identification by MALDI-TOF[34] and antimicrobial susceptibility, as well as novel algorithms for automated reading and interpretation of plates[6][7][8]—could further improve productivity and the quality of results.
Materials and methods
The project
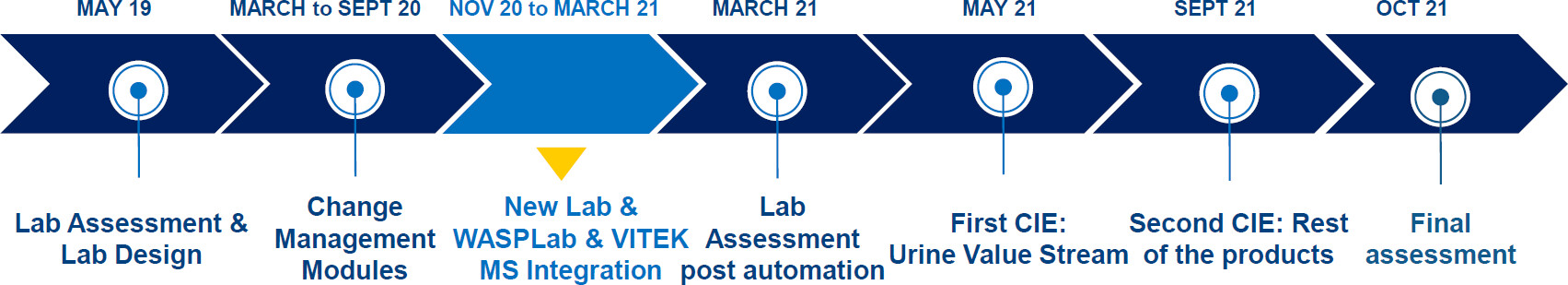
The project took place at the JCS in Lisbon, Portugal between May 2019 and October 2021. In 2019, a full assessment of the laboratory workflow and performance was made to size the instruments’ needs and to propose a lean design for the future microbiology laboratory, considering the different types of workflows of the laboratory. A decision was made to automate the laboratory and remove the UF1000 automated system for urine particle analysis (Sysmex, Japan). The latter move was dictated by the desire to simplify and streamline the processing of this specimen type.
In November 2020, the laboratory moved to the new facilities, where WASPLab (Copan Diagnostics, Inc.) for automated specimen processing and reading and VITEK MS (bioMérieux, France), a MALDI-TOF-based microbial identification system, were installed. For urine samples, a steady routine state was achieved by March 2021. For stool and vaginal swab samples, such a routine was reached by May 2021.
In May 2021, the first CIE event took place and concentrated on the urine workflow. Four months later, in September 2021, a second CIE was conducted for stool and vaginal swab samples. The final assessment of the KPIs was performed in October 2021. The chronology of the project is shown in Fig. 2. Across the three time points, the laboratory processed samples during similar opening times from 8 a.m. to 10 p.m. in two shifts.
|
Pre-conversion state
Laboratory equipment pre-conversion
Before the refurbishment, the lab was equipped with the UF1000 urinalysis system and VITEK 2 system (bioMérieux, France) for microbial identification.
Pre-conversion workflow
The urine sample workflow consisted of many manual steps, from triage to antibiotic sensitivity testing (AST) reporting. Urine samples were screened using the UF1000i (Sysmex, Japan) urinalysis system, which analyzed particles present in the urine by flow cytometry. If the result was suggestive of a positive urine sample, it was streaked onto conventional media and incubated for 24 hours. If the result was suggestive of a negative urine sample, unless a culture was specifically requested by the physician, the sample was discarded without culture. After reading these plates, characteristic colonies were streaked onto CHROMID CPS Elite agar (bioMérieux, France) and incubated for 24 hours. Escherichia coli was identified directly from CHROMID CPS Elite agar; other characteristic colonies were identified using VITEK2. AST was done using VITEK 2 unless other tests were requested.
Vaginal swab samples were received in Stuart medium and cultured on blood agar, Sabouraud, and selective agar containing vancomycin, colistin, amphotericin B, and trimethoprim plates. In case of suspicion of Streptococcus agalactiae, group B was requested, and the sample was streaked onto Strep B agar. Plates were incubated at 35°C–37°C for 48 hours. Identification and AST were done on isolated colonies using VITEK 2 or manual methods.
Stool samples were received in Enteric Transport Medium and then cultured on Hektoen agar incubated for 48 hours at 35°C–37°C, Campylosel agar was incubated for 48 hours at 40°C–42°C, and selenite broth. Pathogens were sub-cultured onto conventional agar before identification and AST using VITEK 2.
Actions taken post-automation
Lean assessment
bioMérieux Lab Consultancy experts applied lean methodology[14][35] to assess the pre- and post-automation situation in the laboratory, including data collection from the LIS, lab organization (e.g., sample volumes and types), staff scheduling, the sample arrival pattern, and positivity rates. Interviews with managers and technologists were performed and direct observation of selected points in the laboratory was carried out. The consultants used snapshots, spaghetti diagrams, volume-vs.-staff analysis, process mapping, time-and-motion study, and brainstorming and problem-solving sessions. During the pre-conversion phase, this information was also used as input to develop the future lean layout of the laboratory capable of accounting for the laboratory’s future needs.
Change management sessions
To best manage the change in operating the JCS laboratory, the following actions were undertaken: assessment to measure the gap between the current practices and the expectations, customization to provide an adapted plan and tuned familiarization modules, and animation to deal with doubts and expectations of staff members and to support the transformation using five hands-on modules based on serious game methodology.
Continuous improvement events
bioMérieux Lab Consultancy experts ran two CIEs (also known as Kaizen events) after WASPLab and VITEK MS reached routine implementation in May and September 2021. CIEs focused on streamlining the workflows around the technology to reduce non-value-added tasks, remove bottlenecks, reduce waiting times, and standardize the non-automated steps. They were delivered following the steps of the A3 thinking lean problem-solving approach (understanding and alignment of the current problem and objectives to achieve, brainstorming and problem-solving to define root causes and potential countermeasures to be implemented, workshops and tests to implement and validate the change, data collection during Kaizen to validate current and future states). The tools applied comprised: 5S (sort, set in order, shine, standardize, sustain), one-piece flow, visual management, and standardized work. The changes implemented after CIEs led to streamlining the workflows based upon the time of sample arrival and standardized reading times per sample type by defining a standard work and displaying priorities on a per hour basis. It was reinforced by implementing a standard huddle one hour before the end of either shift. This corresponded to two five-minute team meetings to review current workload, adapting priorities if needed, and ensuring the standard work was followed. On top of these workflow optimization measures, CIEs led to standardization of bench organization and included the replenishment of benches in the standard work to avoid blockage during the activity peaks. After lab automation, samples were loaded continuously and read after incubation times defined by manufacturers’ recommendations. Characteristic colonies were identified using VITEK MS, and AST was done using VITEK 2 between 8 a.m. and 10 p.m. For urine samples, the standard work included a priority reading between 8 a.m. and 11 a.m. to launch a maximum number of ID and AST analyses before 11 a.m., in order to release a maximum number of results the same day as confirming positivity.
Key performance indicators
Two main KPIs were analyzed: productivity, measured as the number of samples tested per FTE/day, and the turnaround time, defined as the time duration between sample receipt and the validation of all the microbiological documentation, including AST and expressed as median and Q1–Q3 IQR. FTE corresponded to the employee’s scheduled hours divided by the employer’s hours for a full-time workweek. KPIs were measured for negative/positive urine, vaginal swab, and stool samples at three stages of the project, i.e., pre-conversion, post-automation, and post-automation with CIEs. JCS management defined KPIs to be reached following the automation and CIEs, after which, productivity of 100 samples/FTE/day was to be achieved. Desired TATs were defined at 20 hours for negative and 40 hours for positive urine samples, at 45 hours for negative and 60 hours for positive vaginal swab samples, and at 50 hours for negative and 65 hours for positive stool samples (Table 1).
Statistical analysis
Because each period assessed was associated with large and multifactorial technological and organizational changes, median TATs were compared at consecutive time points using Mood’s median test, which does not assume any similarity or difference in group distribution shape. P-values of ≤0.05 were considered to indicate significant differences.
Acknowledgements
Author contributions
This manuscript and its first draft were written by Alicja M. Gruszka, MD, PhD, on behalf of TransPerfect. The medical writing assistance was funded by bioMérieux. The bioMérieux Lab Consultancy and Global Medical Affairs teams participated in data collection, analysis, and review and editing of the manuscript.
G.T.: Investigation, Validation, Writing - review and editing; C.O.: Investigation, Validation, Writing - review and editing; A.R.: Investigation, Validation, Writing - review and editing; S.S.: Investigation, Writing - review and editing; R.P.: Supervision, Writing - review and editing; Y.B.: Validation, Visualization, Writing - review and editing; M.G.G.: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Supervision, Visualization, Writing - review and editing; S.V.: Conceptualization, Methodology, Project administration, Writing - review and editing; V.G.: Investigation, Methodology, Writing - review and editing; C.C.: Conceptualization, Supervision, Writing - review and editing
Funding
This study was supported by bioMérieux, who provides consultancy services and expertise in lab design. No direct funds were provided to Dr. Joaquim Chaves’ laboratory.
Competing interests
G.T., C.O., A.R., S.S., R.P., and C.C. have nothing to disclose. Y.B., M.G.G., S.V., and V.G. are employees of bioMérieux SA. They did not make funding decisions but provided technical support to this project.
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Bailey, Adam L; Ledeboer, Nathan; Burnham, Carey-Ann D (1 May 2019). "Clinical Microbiology Is Growing Up: The Total Laboratory Automation Revolution" (in en). Clinical Chemistry 65 (5): 634–643. doi:10.1373/clinchem.2017.274522. ISSN 0009-9147. https://academic.oup.com/clinchem/article/65/5/634/5608051.
- ↑ 2.0 2.1 2.2 2.3 Cherkaoui, Abdessalam; Schrenzel, Jacques (3 February 2022). "Total Laboratory Automation for Rapid Detection and Identification of Microorganisms and Their Antimicrobial Resistance Profiles". Frontiers in Cellular and Infection Microbiology 12: 807668. doi:10.3389/fcimb.2022.807668. ISSN 2235-2988. PMC PMC8851030. PMID 35186794. https://www.frontiersin.org/articles/10.3389/fcimb.2022.807668/full.
- ↑ Bourbeau, Paul P.; Ledeboer, Nathan A. (1 June 2013). Doern, Gary V.. ed. "Automation in Clinical Microbiology" (in en). Journal of Clinical Microbiology 51 (6): 1658–1665. doi:10.1128/JCM.00301-13. ISSN 0095-1137. PMC PMC3716104. PMID 23515547. https://journals.asm.org/doi/10.1128/JCM.00301-13.
- ↑ 4.0 4.1 4.2 Culbreath, Karissa; Piwonka, Heather; Korver, John; Noorbakhsh, Mir (18 February 2021). McElvania, Erin. ed. "Benefits Derived from Full Laboratory Automation in Microbiology: a Tale of Four Laboratories" (in en). Journal of Clinical Microbiology 59 (3): e01969–20. doi:10.1128/JCM.01969-20. ISSN 0095-1137. PMC PMC8106725. PMID 33239383. https://journals.asm.org/doi/10.1128/JCM.01969-20.
- ↑ Maurer, Florian P.; Christner, Martin; Hentschke, Moritz; Rohde, Holger (30 March 2017). "Advances in Rapid Identification and Susceptibility Testing of Bacteria in the Clinical Microbiology Laboratory: Implications for Patient Care and Antimicrobial Stewardship Programs" (in en). Infectious Disease Reports 9 (1): 6839. doi:10.4081/idr.2017.6839. ISSN 2036-7449. PMC PMC5391540. PMID 28458798. https://www.mdpi.com/2036-7449/9/1/6839.
- ↑ 6.0 6.1 Baker, Justin; Timm, Karen; Faron, Matthew; Ledeboer, Nathan; Culbreath, Karissa (17 December 2020). Munson, Erik. ed. "Digital Image Analysis for the Detection of Group B Streptococcus from ChromID Strepto B Medium Using PhenoMatrix Algorithms" (in en). Journal of Clinical Microbiology 59 (1): e01902–19. doi:10.1128/JCM.01902-19. ISSN 0095-1137. PMC PMC7771474. PMID 33087433. https://journals.asm.org/doi/10.1128/JCM.01902-19.
- ↑ 7.0 7.1 Faron, Matthew L.; Buchan, Blake W.; Relich, Ryan F.; Clark, James; Ledeboer, Nathan A. (25 March 2020). Carroll, Karen C.. ed. "Evaluation of the WASPLab Segregation Software To Automatically Analyze Urine Cultures Using Routine Blood and MacConkey Agars" (in en). Journal of Clinical Microbiology 58 (4): e01683–19. doi:10.1128/JCM.01683-19. ISSN 0095-1137. PMC PMC7098764. PMID 31941690. https://journals.asm.org/doi/10.1128/JCM.01683-19.
- ↑ 8.0 8.1 Faron, Matthew L.; Buchan, Blake W.; Samra, Hasan; Ledeboer, Nathan A. (23 December 2019). Burnham, Carey-Ann D.. ed. "Evaluation of WASPLab Software To Automatically Read chromID CPS Elite Agar for Reporting of Urine Cultures" (in en). Journal of Clinical Microbiology 58 (1): e00540–19. doi:10.1128/JCM.00540-19. ISSN 0095-1137. PMC PMC6935927. PMID 31694967. https://journals.asm.org/doi/10.1128/JCM.00540-19.
- ↑ Antonios, Kritikos; Croxatto, Antony; Culbreath, Karissa (30 December 2021). "Current State of Laboratory Automation in Clinical Microbiology Laboratory" (in en). Clinical Chemistry 68 (1): 99–114. doi:10.1093/clinchem/hvab242. ISSN 0009-9147. https://academic.oup.com/clinchem/article/68/1/99/6490228.
- ↑ Lippi, Giuseppe; Da Rin, Giorgio (27 May 2019). "Advantages and limitations of total laboratory automation: a personal overview" (in en). Clinical Chemistry and Laboratory Medicine (CCLM) 57 (6): 802–811. doi:10.1515/cclm-2018-1323. ISSN 1437-4331. https://www.degruyter.com/document/doi/10.1515/cclm-2018-1323/html.
- ↑ 11.0 11.1 Burckhardt, Irene (22 November 2018). "Laboratory Automation in Clinical Microbiology" (in en). Bioengineering 5 (4): 102. doi:10.3390/bioengineering5040102. ISSN 2306-5354. PMC PMC6315553. PMID 30467275. http://www.mdpi.com/2306-5354/5/4/102.
- ↑ 12.0 12.1 Gibbs, Kellie (30 October 2014), Garcia, Lynne S.; Bachner, Paul; Baselski, Vickie S. et al.., eds., "Managing Change" (in en), Clinical Laboratory Management (Washington, DC, USA: ASM Press): 281–291, doi:10.1128/9781555817282.ch14, ISBN 978-1-68367-106-0, http://doi.wiley.com/10.1128/9781555817282.ch14. Retrieved 2024-03-26
- ↑ Johnson, Pauline M.; Patterson, Claire J.; O'Connell, Mary P. (10 December 2013). "Lean methodology: An evidence-based practice approach for healthcare improvement" (in en). The Nurse Practitioner 38 (12): 1–7. doi:10.1097/01.NPR.0000437576.14143.b9. ISSN 0361-1817. https://journals.lww.com/00006205-201312000-00012.
- ↑ 14.0 14.1 Clark, David M; Silvester, Kate; Knowles, Simon (1 August 2013). "Lean management systems: creating a culture of continuous quality improvement" (in en). Journal of Clinical Pathology 66 (8): 638–643. doi:10.1136/jclinpath-2013-201553. ISSN 0021-9746. http://jcp.bmj.com/lookup/doi/10.1136/jclinpath-2013-201553.
- ↑ Bailey, Adam L.; Burnham, Carey-Ann D. (1 June 2019). "Reducing the time between inoculation and first-read of urine cultures using total lab automation significantly reduces turn-around-time of positive culture results with minimal loss of first-read sensitivity" (in en). European Journal of Clinical Microbiology & Infectious Diseases 38 (6): 1135–1141. doi:10.1007/s10096-019-03512-3. ISSN 0934-9723. http://link.springer.com/10.1007/s10096-019-03512-3.
- ↑ Choi, Qute; Kim, Hyun Jin; Kim, Jong Wan; Kwon, Gye Cheol; Koo, Sun Hoe (1 June 2018). "Manual versus automated streaking system in clinical microbiology laboratory: Performance evaluation of Previ Isola for blood culture and body fluid samples" (in en). Journal of Clinical Laboratory Analysis 32 (5): e22373. doi:10.1002/jcla.22373. ISSN 0887-8013. PMC PMC6817097. PMID 29314254. https://onlinelibrary.wiley.com/doi/10.1002/jcla.22373.
- ↑ Croxatto, Antony; Dijkstra, Klaas; Prod'hom, Guy; Greub, Gilbert (1 July 2015). Burnham, C. -A. D.. ed. "Comparison of Inoculation with the InoqulA and WASP Automated Systems with Manual Inoculation" (in en). Journal of Clinical Microbiology 53 (7): 2298–2307. doi:10.1128/JCM.03076-14. ISSN 0095-1137. PMC PMC4473203. PMID 25972424. https://journals.asm.org/doi/10.1128/JCM.03076-14.
- ↑ Froment, P.; Marchandin, H.; Vande Perre, P.; Lamy, B. (1 March 2014). Diekema, D. J.. ed. "Automated versus Manual Sample Inoculations in Routine Clinical Microbiology: a Performance Evaluation of the Fully Automated InoqulA Instrument" (in en). Journal of Clinical Microbiology 52 (3): 796–802. doi:10.1128/JCM.02341-13. ISSN 0095-1137. PMC PMC3957760. PMID 24353001. https://journals.asm.org/doi/10.1128/JCM.02341-13.
- ↑ Mischnik, Alexander; Mieth, Markus; Busch, Cornelius J.; Hofer, Stefan; Zimmermann, Stefan (1 August 2012). "First Evaluation of Automated Specimen Inoculation for Wound Swab Samples by Use of the Previ Isola System Compared to Manual Inoculation in a Routine Laboratory: Finding a Cost-Effective and Accurate Approach" (in en). Journal of Clinical Microbiology 50 (8): 2732–2736. doi:10.1128/JCM.05501-11. ISSN 0095-1137. PMC PMC3421514. PMID 22692745. https://journals.asm.org/doi/10.1128/JCM.05501-11.
- ↑ Quiblier, Chantal; Jetter, Marion; Rominski, Mark; Mouttet, Forouhar; Böttger, Erik C.; Keller, Peter M.; Hombach, Michael (1 March 2016). Onderdonk, A. B.. ed. "Performance of Copan WASP for Routine Urine Microbiology" (in en). Journal of Clinical Microbiology 54 (3): 585–592. doi:10.1128/JCM.02577-15. ISSN 0095-1137. PMC PMC4767997. PMID 26677255. https://journals.asm.org/doi/10.1128/JCM.02577-15.
- ↑ Graham, Maryza; Tilson, Leanne; Streitberg, Richard; Hamblin, John; Korman, Tony M. (1 September 2016). "Improved standardization and potential for shortened time to results with BD Kiestra™ total laboratory automation of early urine cultures: A prospective comparison with manual processing" (in en). Diagnostic Microbiology and Infectious Disease 86 (1): 1–4. doi:10.1016/j.diagmicrobio.2016.06.020. https://linkinghub.elsevier.com/retrieve/pii/S0732889316301936.
- ↑ Strauss, Sharon; Bourbeau, Paul P. (1 May 2015). Forbes, B. A.. ed. "Impact of Introduction of the BD Kiestra InoqulA on Urine Culture Results in a Hospital Clinical Microbiology Laboratory" (in en). Journal of Clinical Microbiology 53 (5): 1736–1740. doi:10.1128/JCM.00417-15. ISSN 0095-1137. PMC PMC4400793. PMID 25740766. https://journals.asm.org/doi/10.1128/JCM.00417-15.
- ↑ Theparee, Talent; Das, Sanchita; Thomson, Richard B. (1 January 2018). Patel, Robin. ed. "Total Laboratory Automation and Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry Improve Turnaround Times in the Clinical Microbiology Laboratory: a Retrospective Analysis" (in en). Journal of Clinical Microbiology 56 (1): e01242–17. doi:10.1128/JCM.01242-17. ISSN 0095-1137. PMC PMC5744220. PMID 29118171. https://journals.asm.org/doi/10.1128/JCM.01242-17.
- ↑ Mutters, Nico T.; Hodiamont, Caspar J.; Jong, Menno D. de; Overmeijer, Hendri P. J.; Boogaard, Mandy van den; E.Visser, Caroline (1 March 2014). "Performance of Kiestra Total Laboratory Automation Combined with MS in Clinical Microbiology Practice" (in en). Annals of Laboratory Medicine 34 (2): 111–117. doi:10.3343/alm.2014.34.2.111. ISSN 2234-3806. PMC PMC3948823. PMID 24624346. http://annlabmed.org/journal/view.html?doi=10.3343/alm.2014.34.2.111.
- ↑ Da Rin, Giorgio; Zoppelletto, Maira; Lippi, Giuseppe (1 February 2016). "Integration of Diagnostic Microbiology in a Model of Total Laboratory Automation" (in en). Laboratory Medicine 47 (1): 73–82. doi:10.1093/labmed/lmv007. ISSN 0007-5027. https://academic.oup.com/labmed/article-lookup/doi/10.1093/labmed/lmv007.
- ↑ Zhang, Weili; Wu, Siying; Deng, Jin; Liao, Quanfeng; Liu, Ya; Xiong, Li; Shu, Ling; Yuan, Yu et al. (2 December 2021). "Total Laboratory Automation and Three Shifts Reduce Turnaround Time of Cerebrospinal Fluid Culture Results in the Chinese Clinical Microbiology Laboratory". Frontiers in Cellular and Infection Microbiology 11: 765504. doi:10.3389/fcimb.2021.765504. ISSN 2235-2988. PMC PMC8675566. PMID 34926317. https://www.frontiersin.org/articles/10.3389/fcimb.2021.765504/full.
- ↑ Cherkaoui, Abdessalam; Renzi, Gesuele; Martischang, Romain; Harbarth, Stephan; Vuilleumier, Nicolas; Schrenzel, Jacques (28 October 2020). "Impact of Total Laboratory Automation on Turnaround Times for Urine Cultures and Screening Specimens for MRSA, ESBL, and VRE Carriage: Retrospective Comparison With Manual Workflow". Frontiers in Cellular and Infection Microbiology 10: 552122. doi:10.3389/fcimb.2020.552122. ISSN 2235-2988. PMC PMC7664309. PMID 33194794. https://www.frontiersin.org/articles/10.3389/fcimb.2020.552122/full.
- ↑ Yarbrough, Melanie L.; Lainhart, William; McMullen, Allison R.; Anderson, Neil W.; Burnham, Carey-Ann D. (1 December 2018). "Impact of total laboratory automation on workflow and specimen processing time for culture of urine specimens" (in en). European Journal of Clinical Microbiology & Infectious Diseases 37 (12): 2405–2411. doi:10.1007/s10096-018-3391-7. ISSN 0934-9723. http://link.springer.com/10.1007/s10096-018-3391-7.
- ↑ Cornes, Michael P.; Shetty, Chandrashekar (1 June 2018). "Mechanisms of measuring key performance indicators in the pre-analytical phase". Journal of Laboratory and Precision Medicine 3: 57–57. doi:10.21037/jlpm.2018.05.06. http://jlpm.amegroups.com/article/view/4398/5423.
- ↑ Cherkaoui, Abdessalam; Renzi, Gesuele; Viollet, Arnaud; Fleischmann, Mark; Metral-Boffod, Ludovic; Dominguez-Amado, David; Vuilleumier, Nicolas; Schrenzel, Jacques (1 August 2020). "Implementation of the WASPLab™ and first year achievements within a university hospital" (in en). European Journal of Clinical Microbiology & Infectious Diseases 39 (8): 1527–1534. doi:10.1007/s10096-020-03872-1. ISSN 0934-9723. http://link.springer.com/10.1007/s10096-020-03872-1.
- ↑ Leber, Amy L.; Peterson, Ellena; Dien Bard, Jennifer (17 August 2022). Humphries, Romney M.. ed. "The Hidden Crisis in the Times of COVID-19: Critical Shortages of Medical Laboratory Professionals in Clinical Microbiology" (in en). Journal of Clinical Microbiology 60 (8): e00241–22. doi:10.1128/jcm.00241-22. ISSN 0095-1137. PMC PMC9383190. PMID 35658527. https://journals.asm.org/doi/10.1128/jcm.00241-22.
- ↑ Thomson, Richard B.; McElvania, Erin (1 September 2019). "Total Laboratory Automation" (in en). Clinics in Laboratory Medicine 39 (3): 371–389. doi:10.1016/j.cll.2019.05.002. https://linkinghub.elsevier.com/retrieve/pii/S0272271219300290.
- ↑ Cherkaoui, Abdessalam; Schorderet, Didier; Azam, Nouria; Crudeli, Luigi; Fernandez, José; Renzi, Gesuele; Fischer, Adrien; Schrenzel, Jacques (19 October 2022). McElvania, Erin. ed. "Fully Automated EUCAST Rapid Antimicrobial Susceptibility Testing (RAST) from Positive Blood Cultures: Diagnostic Accuracy and Implementation" (in en). Journal of Clinical Microbiology 60 (10): e00898–22. doi:10.1128/jcm.00898-22. ISSN 0095-1137. PMC PMC9580353. PMID 36173195. https://journals.asm.org/doi/10.1128/jcm.00898-22.
- ↑ Heestermans, Robbe; Herroelen, Pauline; Emmerechts, Kristof; Vandoorslaer, Kristof; De Geyter, Deborah; Demuyser, Thomas; Wybo, Ingrid; Piérard, Denis et al. (20 July 2022). Hanson, Kimberly E.. ed. "Validation of the Colibrí Instrument for Automated Preparation of MALDI-TOF MS Targets for Yeast Identification" (in en). Journal of Clinical Microbiology 60 (7): e00237–22. doi:10.1128/jcm.00237-22. ISSN 0095-1137. PMC PMC9297811. PMID 35703578. https://journals.asm.org/doi/10.1128/jcm.00237-22.
- ↑ Novis, David A. (1 September 2008). "Reducing Errors in the Clinical Laboratory: A Lean Production System Approach" (in en). Laboratory Medicine 39 (9): 521–529. doi:10.1309/XAER260DK5U9GM0W. ISSN 1943-7730. https://academic.oup.com/labmed/article-lookup/doi/10.1309/XAER260DK5U9GM0W.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added.