Difference between revisions of "Journal:A metadata-driven approach to data repository design"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) |
||
| Line 57: | Line 57: | ||
: Basing the repository design on collections also reflects the manner in which much modern science is conducted, often via multi-disciplinary collaborations in which each group can generate its own data collections. Collections also greatly facilitate data citation in journal articles. For example, the persistent identifier (DOI) of just the highest collection level of datasets associated with an article can be therein cited, avoiding citation blight. If a particular object (a molecule in our case) is being discussed in the text of the article, it might nevertheless be more appropriate to reference the specific DOI at that stage. Individual citation is also useful in, for example, tables of results or figures. The metadata for any individual cited dataset will also contain the attribute "is member of," so that the hierarchy can be both tracked upwards, and via the attribute "has members" downwards (Fig. 1d). This hierarchy also introduces via such metadata further semantics into the citation process itself; each item is placed into appropriate context. Lack of such semantics/context are arguably one of the most deficient aspects of current citation practices in journal articles. | : Basing the repository design on collections also reflects the manner in which much modern science is conducted, often via multi-disciplinary collaborations in which each group can generate its own data collections. Collections also greatly facilitate data citation in journal articles. For example, the persistent identifier (DOI) of just the highest collection level of datasets associated with an article can be therein cited, avoiding citation blight. If a particular object (a molecule in our case) is being discussed in the text of the article, it might nevertheless be more appropriate to reference the specific DOI at that stage. Individual citation is also useful in, for example, tables of results or figures. The metadata for any individual cited dataset will also contain the attribute "is member of," so that the hierarchy can be both tracked upwards, and via the attribute "has members" downwards (Fig. 1d). This hierarchy also introduces via such metadata further semantics into the citation process itself; each item is placed into appropriate context. Lack of such semantics/context are arguably one of the most deficient aspects of current citation practices in journal articles. | ||
* Our approach to metadata collection is to automate the process whenever possible. In the case of a molecule as an object, there are algorithms which can be used to generate appropriate metadata, the most useful and prominent of which is the InChI (International Chemical Identifier).<ref name="InChI">{{cite web |url=http://www.inchi-trust.org/ |title=InChI Trust |publisher=InChI Trust |accessdate=07 September 2016}}</ref> The task of creating such an identifier is effectively accomplished using the OpenBabel program library<ref name="OBoyleOpen11">{{cite journal |title=Open Babel: An open chemical toolbox |journal=Journal of Cheminformatics |author=O'Boyle, N.M.; Banck, M.; James, C.A. et al. |volume=3 |pages=33 |year=2011 |doi=10.1186/1758-2946-3-33 |pmid=21982300 |pmc=PMC3198950}}</ref> or via Javascript-based resources.<ref name="InChI.js">{{cite web |url=https://metamolecular.com/inchi-js/ |title=InChI for the Web Browser with InChI.js |publisher=Metamolecular, LLC |accessdate=07 September 2016}}</ref> These can accept as input a variety of chemical documents and generate an appropriate InChI identifier and InChI key uniquely describing them. The repository workflow automatically processes any uploaded data file through this algorithm and records all successful outputs. Such metadata is then associated with the Subject element in the DataCite schema (Fig. 1b). | * Our approach to metadata collection is to automate the process whenever possible. In the case of a molecule as an object, there are algorithms which can be used to generate appropriate metadata, the most useful and prominent of which is the InChI (International Chemical Identifier).<ref name="InChI">{{cite web |url=http://www.inchi-trust.org/ |title=InChI Trust |publisher=InChI Trust |accessdate=07 September 2016}}</ref> The task of creating such an identifier is effectively accomplished using the OpenBabel program library<ref name="OBoyleOpen11">{{cite journal |title=Open Babel: An open chemical toolbox |journal=Journal of Cheminformatics |author=O'Boyle, N.M.; Banck, M.; James, C.A. et al. |volume=3 |pages=33 |year=2011 |doi=10.1186/1758-2946-3-33 |pmid=21982300 |pmc=PMC3198950}}</ref> or via Javascript-based resources.<ref name="InChI.js">{{cite web |url=https://metamolecular.com/inchi-js/ |title=InChI for the Web Browser with InChI.js |publisher=Metamolecular, LLC |accessdate=07 September 2016}}</ref> These can accept as input a variety of chemical documents and generate an appropriate InChI identifier and InChI key uniquely describing them. The repository workflow automatically processes any uploaded data file through this algorithm and records all successful outputs. Such metadata is then associated with the <tt>Subject</tt> element in the DataCite schema (Fig. 1b). | ||
==References== | ==References== | ||
Revision as of 22:29, 7 February 2017
| Full article title | A metadata-driven approach to data repository design |
|---|---|
| Journal | Journal of Cheminformatics |
| Author(s) | Harvey, Matthew J.; McLean, Andrew; Rzepa, Henry S. |
| Author affiliation(s) | Imperial College London |
| Primary contact | Email: rzepa at imperial dot ac dot uk |
| Year published | 2017 |
| Volume and issue | 9 |
| Page(s) | 4 |
| DOI | 10.1186/s13321-017-0190-6 |
| ISSN | 1758-2946 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | http://jcheminf.springeropen.com/articles/10.1186/s13321-017-0190-6 |
| Download | http://jcheminf.springeropen.com/track/pdf/10.1186/s13321-017-0190-6 (PDF) |
|
|
This article should not be considered complete until this message box has been removed. This is a work in progress. |
Abstract
The design and use of a metadata-driven data repository for research data management is described. Metadata is collected automatically during the submission process whenever possible and is registered with DataCite in accordance with their current metadata schema, in exchange for a persistent digital object identifier. Two examples of data preview are illustrated, including the demonstration of a method for integration with commercial software that confers rich domain-specific data analytics without introducing customization into the repository itself.
Keywords: Data repository, metadata-driven, DataCite, data preview, Mpublish
Background
Turnkey institutional repositories based on platforms such as DSpace[1] were introduced more than 10 years ago, with early applications directed largely towards archival of publication preprints and postprints. The recent increasing requirement for research data management emerging from funding agencies means that the focus is now shifting to the use of repositories as part of the data management processes. More recent data-centric tools such as Figshare[2] and Zenodo[3] reflect these changes. Such services rely on the minting of persistent identifiers or DOIs for the depositions using the DataCite agency.[4] Metadata describing the deposited material is supplied to DataCite and a DOI is returned. An early example of such research data management is illustrated by a DSpace-based project to produce and, 10 years later, curate a library of quantum-mechanically-optimised molecular coordinates derived from a computable subset of the National Cancer Institute's (NCI) collection of small molecules.[5]
One feature of the curation phase[6] of the project aimed to explore the capabilities of the DataCite metadata schemas to improve the discoverability of the deposited data. The metadata can then be exploited to create rich search queries.[7] As a result of the experiences gained from this project, we became aware that one limiting factor to the effective use of metadata was the repository design itself. The next stage therefore was to explore whether what we considered the essential requirements for a data repository could be incorporated into a new design. Here we report the principles used to create such a repository and some of the applications in chemistry that have resulted. These principles may in turn assist researchers wishing to deposit data in identifying the repository attributes that can best expose the discoverability and re-use of their data.
Data repository design features
Here we describe the requirements we identified for a metadata-driven repository, an instance of which is deployed by the Imperial College HPC Service at https://data.hpc.imperial.ac.uk:
- In our design, we have focused on enhancing the FAIR[8] attributes of the data. The first attribute F means the data must be findable and practically this means making the metadata descriptors as rich and complete as possible to enable this. A = Accessibility is achieved by assigning persistent identifiers to the datasets and again associating them with appropriate metadata to enable automated retrieval processes if appropriate. This in turn helps ensure that the data can be accessed in a standard manner to enable its inter-operability in various software environments. R = Re-usability is related to understanding and trusting its provenance and the license terms under which it can be processed.
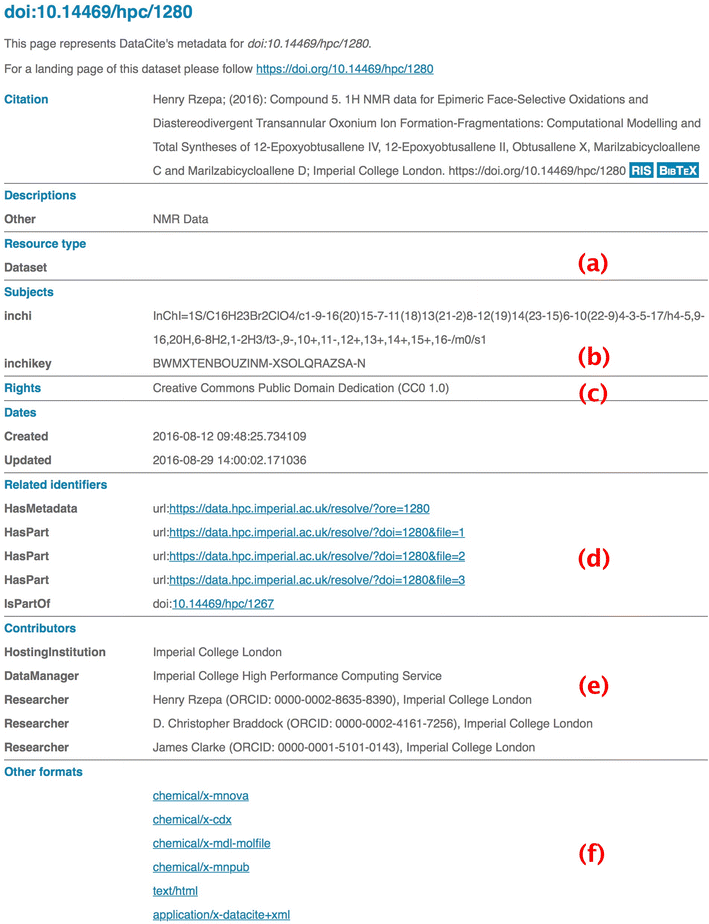
- The provenance of the deposited data is established from the unique ORCiD identifier of the depositor(s). On the first occasion that the repository is used after initial institutional-based authentication, a redirection to the ORCiD site occurs. There the depositor creates an account or authenticates an existing account, followed by authorising the repository request. The retrieved ORCiD is then added to the metadata manifest for the deposition as a depositor attribute. This initial depositor can then add further ORCiDs as co-authors to the entry; these again are validated automatically from the ORCiD site. This information is then collected and sent to DataCite for aggregation (Fig. 1e).
|
- The structure of the repository is based on hierarchical collections. Although collections have been a feature of early repositories such as DSpace, relatively little use has been made of them. We first identified the need for such structures from our early project[5] involving individual deposition of >168,000 items. This was deemed necessary since we considered that each item would benefit from having its own unique metadata descriptors, but within the context of a complete collection described using separate metadata. This is illustrated by assigning metadata both to individual entries[9] and to the collection which the individual items are members of.[10] Such hierarchical structures allow a research group to assign collections to project themes and within these to identify sub-collections associated with individual researchers or teams. The sub-collections can be further structured into types of data, other research objects such as software, presentations on the topic and other media such as video. The granularity of this approach is likely to depend very much on the discipline associated with the data. Thus in molecular sciences, the basic object naturally maps to the molecule, since this is the smallest object for which a dataset can be normally be generated and which can usefully be described by its own metadata. It would be less useful or convenient, for example, to disassemble the molecule into individual atoms as metadata carriers.
- Basing the repository design on collections also reflects the manner in which much modern science is conducted, often via multi-disciplinary collaborations in which each group can generate its own data collections. Collections also greatly facilitate data citation in journal articles. For example, the persistent identifier (DOI) of just the highest collection level of datasets associated with an article can be therein cited, avoiding citation blight. If a particular object (a molecule in our case) is being discussed in the text of the article, it might nevertheless be more appropriate to reference the specific DOI at that stage. Individual citation is also useful in, for example, tables of results or figures. The metadata for any individual cited dataset will also contain the attribute "is member of," so that the hierarchy can be both tracked upwards, and via the attribute "has members" downwards (Fig. 1d). This hierarchy also introduces via such metadata further semantics into the citation process itself; each item is placed into appropriate context. Lack of such semantics/context are arguably one of the most deficient aspects of current citation practices in journal articles.
- Our approach to metadata collection is to automate the process whenever possible. In the case of a molecule as an object, there are algorithms which can be used to generate appropriate metadata, the most useful and prominent of which is the InChI (International Chemical Identifier).[11] The task of creating such an identifier is effectively accomplished using the OpenBabel program library[12] or via Javascript-based resources.[13] These can accept as input a variety of chemical documents and generate an appropriate InChI identifier and InChI key uniquely describing them. The repository workflow automatically processes any uploaded data file through this algorithm and records all successful outputs. Such metadata is then associated with the Subject element in the DataCite schema (Fig. 1b).
References
- ↑ "DSpace". DuraSpace Organization. http://www.dspace.org/. Retrieved 07 September 2016.
- ↑ "Zenodo". CERN Data Centre. https://zenodo.org/. Retrieved 07 September 2016.
- ↑ "DataCite". DataCite Association. https://www.datacite.org/. Retrieved 07 September 2016.
- ↑ 5.0 5.1 Downing, J.; Murray-Rust, P.; Tonge, A.P. et al. (2008). "SPECTRa: The deposition and validation of primary chemistry research data in digital repositories". Journal of Chemical Information and Modeling 48 (8): 1571–1581. doi:10.1021/ci7004737.
- ↑ Harvey, M.J.; Mason, N.J.; McLean, A. et al. (2015). "Standards-based curation of a decade-old digital repository dataset of molecular information". Journal of Cheminformatics 7: 43. doi:10.1186/s13321-015-0093-3. PMC PMC4550659. PMID 26322133. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4550659.
- ↑ Rzepa, H.S.; Mclean, A.; Harvey, M.J. (2015). "InChI as a research data management tool". Chemistry International 38 (3–4): 24–26. doi:10.1515/ci-2016-3-408.
- ↑ Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J. et al. (2016). "The FAIR Guiding Principles for scientific data management and stewardship". Scientific Data 3: 160018. doi:10.1038/sdata.2016.18. PMC PMC4792175. PMID 26978244. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4792175.
- ↑ "doi:10.14469/ch/153690". DataCite Content Service Beta. DataCite Association. https://data.datacite.org/10.14469/ch/153690. Retrieved 07 September 2016.
- ↑ "doi:10.14469/ch/2". DataCite Content Service Beta. DataCite Association. https://data.datacite.org/10.14469/ch/2. Retrieved 07 September 2016.
- ↑ "InChI Trust". InChI Trust. http://www.inchi-trust.org/. Retrieved 07 September 2016.
- ↑ O'Boyle, N.M.; Banck, M.; James, C.A. et al. (2011). "Open Babel: An open chemical toolbox". Journal of Cheminformatics 3: 33. doi:10.1186/1758-2946-3-33. PMC PMC3198950. PMID 21982300. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3198950.
- ↑ "InChI for the Web Browser with InChI.js". Metamolecular, LLC. https://metamolecular.com/inchi-js/. Retrieved 07 September 2016.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. In one case, the original citation was incomplete (#6) and was corrected here.