Difference between revisions of "Journal:Privacy-preserving healthcare informatics: A review"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) |
||
| Line 332: | Line 332: | ||
''l''-diversity requires every QID group to contain at least ''l'' distinct sensitive attribute values. For example, Table 3 is a 3-diverse table where there are at least three distinct sensitive attribute values for every QID group. This method depends on the range of the sensitive attribute values. If the number of distinct sensitive attribute values is lower than the desired privacy parameter ''l'', some fictitious data are added to achieve ''l''-diversity. This further leads to excessive modification and may produce biased results in statistical analysis. In addition, ''l''-diversity does not prevent attribute disclosure when the overall distribution of the sensitive attribute is skewed. A skewness attack and similarity attack are still possible to disclose the SA values in ''l''-diversity. ''k''-anonymity and ''l''-diversity have been combined to propose ''𝜏''-safe (''l'', ''k'')-diversity.<ref name="ZhuSafe18">{{cite journal |title=τ-Safe ( l,k )-Diversity Privacy Model for Sequential Publication With High Utility |journal=IEEE Access |author=Zhu, H.; Liang, H.-B.; Zhao, L. et al. |volume=7 |pages=687–701 |year=2018 |doi=10.1109/ACCESS.2018.2885618}}</ref> | ''l''-diversity requires every QID group to contain at least ''l'' distinct sensitive attribute values. For example, Table 3 is a 3-diverse table where there are at least three distinct sensitive attribute values for every QID group. This method depends on the range of the sensitive attribute values. If the number of distinct sensitive attribute values is lower than the desired privacy parameter ''l'', some fictitious data are added to achieve ''l''-diversity. This further leads to excessive modification and may produce biased results in statistical analysis. In addition, ''l''-diversity does not prevent attribute disclosure when the overall distribution of the sensitive attribute is skewed. A skewness attack and similarity attack are still possible to disclose the SA values in ''l''-diversity. ''k''-anonymity and ''l''-diversity have been combined to propose ''𝜏''-safe (''l'', ''k'')-diversity.<ref name="ZhuSafe18">{{cite journal |title=τ-Safe ( l,k )-Diversity Privacy Model for Sequential Publication With High Utility |journal=IEEE Access |author=Zhu, H.; Liang, H.-B.; Zhao, L. et al. |volume=7 |pages=687–701 |year=2018 |doi=10.1109/ACCESS.2018.2885618}}</ref> | ||
To address these vulnerabilities, ''t''-closeness has been proposed, which requires that the distribution of a sensitive attribute in any equivalence class to be close to the distribution of the attribute in the overall table. That is, the distance between the distributions is less than a threshold. This property prevents an adversary from making an accurate estimation of the sensitive attribute values and thus preventing attribute disclosure. However, only SA values are modified, while all the QID values remain unchanged in this model. Hence, it does not prevent identity disclosure. Furthermore, ''t''-closeness deploys a brute-force approach to examine each possible partition of the table to find the optimal solution. This process takes an enormous computation time complexity of 2<sup>''𝑂''(''𝑛'')''𝑂''(''𝑚'')</sup>. | |||
To address membership disclosure, ''𝛿''-presence has been proposed to limit the confidence level of an adversary in inferring the existence of a targeted victim in the published data to at most ''𝛿''%. | |||
There is a significant amount of precedent for a different parameter value of the privacy models, which could be used as benchmarks for efficient data publishing. However, the choice of the privacy parameter value is flexible and depends on the desired privacy and utility objectives of the data publication, provided that "an acceptable privacy level" is guaranteed. | |||
===Differential privacy=== | |||
Figure 3 shows a data publishing scenario in differential privacy. Differential privacy<ref name="DworkDiffer08">{{cite journal |title=Differential Privacy: A Survey of Results |journal=Proceedings of the 2008 International Conference on Theory and Applications of Models of Computation |author=Dwork, C. |pages=1–19 |year=2008 |doi=10.1007/978-3-540-79228-4_1}}</ref><ref name="AlnemariAnAdapt17">{{cite journal |title=An Adaptive Differential Privacy Algorithm for Range Queries over Healthcare Data |journal=Proceedings of the 2017 IEEE International Conference on Healthcare Informatics |author=Alnemari, S.; Romanowski, C.J.; Raj, R.K. |pages=397–402 |year=2017 |doi=10.1109/ICHI.2017.49}}</ref><ref name="LiEffic15">{{cite journal |title=Efficient e-health data release with consistency guarantee under differential privacy |journal=Proceedings of the 17th International Conference on E-health Networking, Application & Services |author=Li, H.; Dai, Y.; Lin, X. |pages=602–608 |year=2015 |doi=10.1109/HealthCom.2015.7454576}}</ref><ref name="GuttierezUser18">{{cite journal |title=User-Centered Differential Privacy Mechanisms for Electronic Medical Records |journal=Proceedings of the 2018 International Carnahan Conference on Security Technology |author=Guttierrez, O.; Saavedra, J.J.; Zurbaran, M. et al. |pages=1–5 |year=2018 |doi=10.1109/CCST.2018.8585555}}</ref> involves a query answering process, which a data recipient may send a query to the database and the result of that query is probabilistically indistinguishable regardless of the presence of a record in the database. That is, given two databases that differ in exactly one record, a differentially private mechanism provides two randomized outputs that have almost similar probability distributions. In other words, an adversary could not infer the existence of a targeted victim in the published database with high probability. Randomized noise derived from Laplace distribution is added to the result of the query to achieve privacy. | |||
Revision as of 20:40, 11 April 2021
| Full article title | Privacy-preserving healthcare informatics: A review |
|---|---|
| Journal | ITM Web of Conferences |
| Author(s) | Chong, Kah Meng |
| Author affiliation(s) | Universiti Tunku Abdul Rahman |
| Primary contact | kmchong at utar dot edu dot my |
| Year published | 2021 |
| Volume and issue | 36 |
| Article # | 04005 |
| DOI | 10.1051/itmconf/20213604005 |
| ISSN | 2271-2097 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.itm-conferences.org/articles/itmconf/abs/2021/01/itmconf_icmsa2021_04005/ |
| Download | https://www.itm-conferences.org/articles/itmconf/pdf/2021/01/itmconf_icmsa2021_04005.pdf (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
The electronic health record (EHR) is the key to an efficient healthcare service delivery system. The publication of healthcare data is highly beneficial to healthcare industries and government institutions to support a variety of medical and census research. However, healthcare data contains sensitive information of patients, and the publication of such data could lead to unintended privacy disclosures. In this paper, we present a comprehensive survey of the state-of-the-art privacy-enhancing methods that ensure a secure healthcare data sharing environment. We focus on the recently proposed schemes based on data anonymization and differential privacy approaches in the protection of healthcare data privacy. We highlight the strengths and limitations of the two approaches and discuss some promising future research directions in this area.
Keywords: data privacy, data sharing, electronic health record, healthcare informatics,
Introduction
Electronic health record (EHR) systems are increasingly adopted as an important paradigm in the healthcare industry to collect and store patient data, which includes sensitive information such as demographic data, medical history, diagnosis code, medications, treatment plans, hospitalization records, insurance information, immunization dates, allergies, and laboratory and test results. The availability of such big data has provided unprecedented opportunities to improve the efficiency and quality of healthcare services, particularly in improving patient care outcomes and reducing medical costs. EHR data have been published to allow useful analysis as required by the healthcare industry[1] and government institutions.[2][3] Some key examples may include large-scale statistical analytics (e.g., the study of correlation between diseases), clinical decision making, treatment optimization, clustering (e.g., epidemic control), and census surveys. Driven by the potential of EHR systems, a number of EHR repositories have been established, such as the National Database for Autism Research (NDAR), U.K. Data Service, ClinicalTrials.gov, and UNC Health Care (UNCHC).
Although the publication of EHR data is enormously beneficial, it could lead to unintended privacy disclosures. Many conventional cryptography and security methods have been deployed to primarily protect the security of EHR systems, including access control, authentication, and encryption. However, these technologies do not guarantee privacy preservation of sensitive data. That is, the sensitive information of patient could still be inferred from the published data by an adversary. Various regulations and guidelines have been developed to restrict publishable data types, data usage, and data storage, including the Health Insurance Portability and Accountability Act (HIPAA)[4][5], General Data Protection Regulation (GDPR)[6][7], and Personal Data Protection Act.[8] However, there are several limitations to this regulatory approach. First, a high trust level is required of the data recipient that they follow the rules and regulations provided by the data publisher. Yet, there are adversaries who attempt to attack the published data to reidentify a target victim. Second, sensitive data still might be carelessly published due to human error and fall into the wrong hands, which eventually leads to a breach of individual privacy. As such, regulations and guidelines alone do not provide computational guarantee for preserving the privacy of a patient and thus cannot fully prevent such privacy violations. The need of protecting individual data privacy in a hostile environment, while allowing accurate analysis of patient data, has driven the development of effective privacy models in protecting healthcare data.
In this paper, we present the privacy issues in healthcare data publication and elaborate on relevant adversarial attack models. With a focus on data anonymization and differential privacy, we discuss the limitations and strengths of these proposed approaches. Finally, we conclude the paper and highlight future research direction in this area.
Privacy threats
In this section, we first discuss privacy-preserving data publishing (PPDP) and the properties of healthcare data. Then, we present the major privacy disclosures in healthcare data publication and show the relevant attack models. Finally, we present the privacy and utility objective in PPDP.
Privacy-preserving data publishing
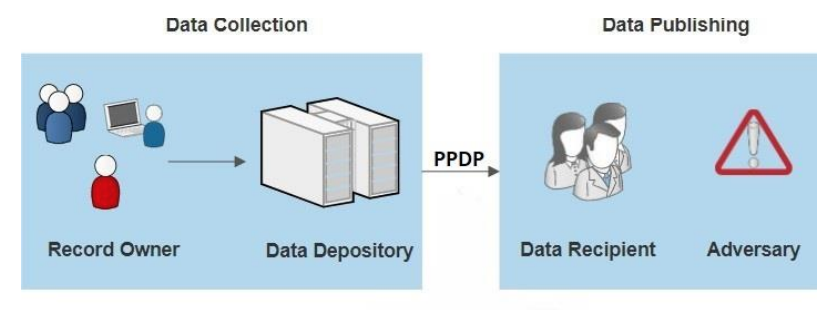
Privacy-preserving data publishing (PPDP) provides technical solutions that address the privacy and utility preservation challenges of data sharing scenarios. An overview of PPDP is shown in Figure 1, which includes a general data collection and data publishing scenario.
|
During the data collection phase, data of the record owner (patient) are collected by the data holder (hospital) and stored in an EHR. In the data publishing phase, the data holder releases the collected data to the data recipient (e.g., the public or a third party, e.g., an insurance company or medical research center) for further analysis and data mining. However, some of the data recipients (adversary) are not honest and attempt to obtain more information about the record owner beyond the published data, which includes the identity and sensitive data of the record owner. Hence, PPDP serves as a vital process that sanitizes sensitive information to avoid privacy violations of one or more individuals.
Healthcare data
Typically, stored healthcare data exists as relational data in tabular form. Each row (tuple) corresponds to one record owner, and each column corresponds to a number of distinct attributes, which can be grouped into the following four categories:
- Explicit identifier (ID): a set of attributes such as name, social security number, national IDs, mobile number, and drivers license number that uniquely identifies a record owner
- Quasi-identifier (QID): a set of attributes such as date of birth, gender, address, zip code, and hobby that cannot uniquely identify a record owner but can potentially identify the target if combined with some auxiliary information
- Sensitive attribute (SA): sensitive personal information such as diagnosis codes, genomic information, salary, health condition, insurance information, and relationship status that the record owner intends to keep private from unauthorized parties
- Non-sensitive attribute (NSA): a set of attributes such as cookie IDs, hashed email addresses, and mobile advertising IDs generated from an EHR that do not violate the privacy of the record owner if they are disclosed (Note: all attributes that are not categorized as ID, QID, and SA are classified as NSA.)
Each attribute can be further classified as a numerical attribute (e.g., age, zip code, and date of birth) and non-numerical attribute (e.g., gender, job, and disease). Table 1 shows an example dataset, in which the name of patients is naively anonymized (by removing the names and social security numbers).
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Privacy disclosures
A privacy disclosure is defined as a disclosure of personal information that users intend to keep private from an entity which is not authorized to access or have the information. There are three types of privacy disclosures:
- Identity disclosure: Identity disclosure, also known as reidentification, is the major privacy threat in publishing healthcare data. It occurs when the true identity of a targeted victim is revealed by an adversary from the published data. In other words, an individual is reidentified when an adversary is able to map a record in the published data to its corresponding patient with high probability (record linkage). For example, if an adversary possesses the information that A is 43 years old, then A is reidentified as record 7 in Table 1.
- Attribute disclosure: This disclosure occurs when an adversary successfully links a victim to their SA information in the published data with high probability (attribute linkage). This SA information could be an SA value (e.g., "Disease" in Table 1) or a range that contains the SA value (e.g., medical cost range).
- Membership disclosure: This disclosure occurs when an adversary successfully infers the existence of a targeted victim in the published data with high probability. For example, the inference of an individual in a COVID-19-positive database poses a privacy threat to the individual.
Attack models
Privacy attacks could be launched by matching a published table containing sensitive information about the target victim with some external resources modelling the background knowledge of the attacker. For a successful attack, an adversary may require the following prior knowledge:
- The published table, 𝑻′: An adversary has access to the published table 𝑇′ (which is often an open resource) and knows that 𝑇 is an anonymized data for some table T.
- QID of a targeted victim: An adversary possesses partial or complete QID values about a target from any external resource and the values are accurate. This assumption is realistic as the QID information is easy to acquire from different sources, including real-life inspection data, external demographic data, and and voter list data.
- Knowledge about the distribution of the SA and NSA in table T : For example, an adversary may possess the information of P (disease=diabetes, age>50) and may utilize this knowledge to make additional inferences about records in the published table 𝑇′.
Generally, privacy attacks could be launched due to the linkability properties of the QID. Now we discuss the relevant privacy attack models for identity and attribute disclosure.
- Linkage attack: An adversary may reidentify the identity and discover the SA values of a targeted record owner by matching the auxiliary QID values with the published table 𝑇′. For example, imagine Table 1 is published without modification and suppose A possesses the knowledge that B lives in zip code 96038, then A infers that B belongs to record 1 (identity disclosure) and has diabetes (attribute disclosure).[9][10][11][12][13]
- Homogeneity attack: This attack discloses the SA values of a target when there is insufficient homogeneity in the SA. That is, the combination of QID is mapped to one SA value only. For example, suppose A knows that B is 28 years old, which belongs to the first equivalence class (an equivalence class is a cluster of records with the same QID values) in Table 2, below (record 1, 2 and 3). Since these records have the same disease, A infers that B suffers from diabetes.[9][14]
- Background knowledge attack: This attack utilizes logical reasoning and additional knowledge about a target to breach the SA values. For example, suppose A knows that C is 43 years old and lives in the zip code 96583, which belongs to the third equivalence class in Table 2, below (record 7, 8 and 9). Nevertheless, the records show that C may have either diabetes or cancer. Based on A’s background knowledge that C is a person who likes sweet foods, A infers that C is diabetic.[9][14]
- Skewness attack: When the overall distribution of SA in the original data is skewed, SA values can be inferred. The SA values have different degrees of sensitivity. For instance, a victim may not mind being known as diabetic as it is a common (majority) disease. However, one would mind being known to have mental illness. According to Table 3, below, the probability of having mental illness is 33.3%, which is much higher than that of real distribution (11.1% in Table 1). Thus, this imposes a privacy threat, since anyone in the equivalence class that has a 33.3% possibility can be inferred to have mental illness, as compared with 11.1% of the overall distribution.[15]
- Similarity attack: This attack discloses SA values when the semantic relationship of distinct SA values in an equivalence class is close. For example, suppose that an adversary infers the possible salaries of a target victim are 2K, 3K, and 4K. Although the numbers represent distinct salary, they are all categorized in the range [2K,4K]. Hence, an adversary could infer that the target has low salary when the SA values are semantically similar.[14][15]
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Privacy and utility objective of PPDP
PPDP allows computational guarantees on the prevention of privacy disclosures while maintaining the usefulness of the published data. From the privacy aspect, the identity of patients and their corresponding SA values should be concealed from the public. For instance, it is permissible to disclose the information that there exists diabetic patients in the hospital, but the published data should not disclose which patients have diabetes. Utility preservation is another aspect of PPDP, which emphasizes publishing data that is “almost similar” to the original data. Given that M is an arbitrary data mining process, the output of M(T) and M(𝑇′) should be almost similar: the difference between M(T) and M(𝑇′) should be less than a threshold t. In most PPDP scenarios, the data mining process M (the usage of the published data) is unknown at the time of publication. This process M could be a simple census statistic or some specified analysis and data exploration, such as pattern mining, association rules, and data modelling. Privacy and utility are two contradictory aspects: publishing high-utility data implies less privacy protection to the record owner and vice versa.
Privacy models
In this section, we present some well-established privacy models that are used to ensure privacy in healthcare data. Particularly, we focus on data anonymization and differential privacy as two mainstream PPDP technologies that are different in their data publishing mechanisms.
Data anonymization
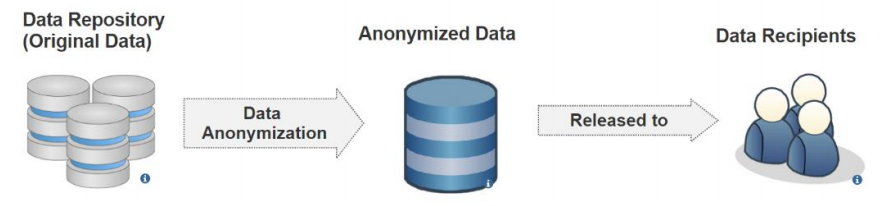
Figure 2 shows a data publishing scenario in data anonymization. An original database is modified before being published as an anonymized database, which is generated by deploying generalization and suppression on the original database. The anonymized database could be studied in place of the original database. Some common data anonymization models to prevent privacy disclosure include k-anonymity[10][11][12][13][14], l-diversity[9], t-closeness[15], and 𝛿-presence.[16]
|
k-anonymity was developed to address identity disclosure. It requires that, for one record in the table that has some QID value, there exists at least k-1 other records in the table that have the same QID value. Hence, each record is indistinguishable from at least k-1 other records with respect to the QID value in a k-anonymous table. For example, Table 2 and 3 are 3-anonymous tables. In k-anonymity, any individual cannot be reidentified from the published data with a probability of higher than 1/k. Other variations of k-anonymity include clustering anonymity[11], distribution-preserving k-anonymity[12], optimization-based k-anonymity[13], 𝜃'-sensitive k-anonymity[14], (X,Y)-anonymity[17], (α, k)-anonymity[18], LKC-privacy[19], and random k-anonymous[20], which prevents identity disclosure by hiding the record of a target in an equivalence class of records with the same QID values. Although the k-anonymity model protects against identity disclosure, it is vulnerable against attribute disclosure. A homogeneity attack and background knowledge attack is possible by deducing the sensitive attribute values from the published data. To provide protection on the sensitive attribute value, l-diversity and t-closeness have been proposed.
l-diversity requires every QID group to contain at least l distinct sensitive attribute values. For example, Table 3 is a 3-diverse table where there are at least three distinct sensitive attribute values for every QID group. This method depends on the range of the sensitive attribute values. If the number of distinct sensitive attribute values is lower than the desired privacy parameter l, some fictitious data are added to achieve l-diversity. This further leads to excessive modification and may produce biased results in statistical analysis. In addition, l-diversity does not prevent attribute disclosure when the overall distribution of the sensitive attribute is skewed. A skewness attack and similarity attack are still possible to disclose the SA values in l-diversity. k-anonymity and l-diversity have been combined to propose 𝜏-safe (l, k)-diversity.[21]
To address these vulnerabilities, t-closeness has been proposed, which requires that the distribution of a sensitive attribute in any equivalence class to be close to the distribution of the attribute in the overall table. That is, the distance between the distributions is less than a threshold. This property prevents an adversary from making an accurate estimation of the sensitive attribute values and thus preventing attribute disclosure. However, only SA values are modified, while all the QID values remain unchanged in this model. Hence, it does not prevent identity disclosure. Furthermore, t-closeness deploys a brute-force approach to examine each possible partition of the table to find the optimal solution. This process takes an enormous computation time complexity of 2𝑂(𝑛)𝑂(𝑚).
To address membership disclosure, 𝛿-presence has been proposed to limit the confidence level of an adversary in inferring the existence of a targeted victim in the published data to at most 𝛿%.
There is a significant amount of precedent for a different parameter value of the privacy models, which could be used as benchmarks for efficient data publishing. However, the choice of the privacy parameter value is flexible and depends on the desired privacy and utility objectives of the data publication, provided that "an acceptable privacy level" is guaranteed.
Differential privacy
Figure 3 shows a data publishing scenario in differential privacy. Differential privacy[22][23][24][25] involves a query answering process, which a data recipient may send a query to the database and the result of that query is probabilistically indistinguishable regardless of the presence of a record in the database. That is, given two databases that differ in exactly one record, a differentially private mechanism provides two randomized outputs that have almost similar probability distributions. In other words, an adversary could not infer the existence of a targeted victim in the published database with high probability. Randomized noise derived from Laplace distribution is added to the result of the query to achieve privacy.
References
- ↑ Senthilkumar, S.A.; Rai, B.K.; Meshram, A.A. et al. (2018). "Big Data in Healthcare Management: A Review of Literature". American Journal of Theoretical and Applied Business 4 (2): 57–69. doi:10.11648/j.ajtab.20180402.14.
- ↑ Dudeck, M.A.; Horan, T.C.; Peterson, K.D. et al. (2011). "National Healthcare Safety Network (NHSN) Report, data summary for 2010, device-associated module". American Journal of Infection Control 39 (10): 798-816. doi:10.1016/j.ajic.2011.10.001. PMID 22133532.
- ↑ Powell, K.M.; Li, Q.; Gross, C. et al. (2019). "Ventilator-Associated Events Reported by U.S. Hospitals to the National Healthcare Safety Network, 2015-2017". Proceedings of the American Thoracic Society 2019 International Conference. doi:10.1164/ajrccm-conference.2019.199.1_MeetingAbstracts.A3419.
- ↑ Cohen, I.G.; Mello, M.M. (2018). "HIPAA and Protecting Health Information in the 21st Century". JAMA 320 (3): 231–32. doi:10.1001/jama.2018.5630. PMID 29800120.
- ↑ Obeng, O.; Paul, S. (2019). "Understanding HIPAA Compliance Practice in Healthcare Organizations in a Cultural Context". AMCIS 2019 Proceedings: 1–5. https://aisel.aisnet.org/amcis2019/info_security_privacy/info_security_privacy/1/.
- ↑ Voigt, P.; von dem Bussche, A. (2017). The EU General Data Protection Regulation (GDPR): A Practical Guide. Springer. ISBN 9783319579580.
- ↑ Tikkinen-Piri, C.; Rohunen, A.; Markkula, J. (2018). "EU General Data Protection Regulation: Changes and implications for personal data collecting companies". Computer Law & Security Review 34 (1): 134–53. doi:10.1016/j.clsr.2017.05.015.
- ↑ Carey, P. (2018). Data Protection: A Practical Guide to UK and EU Law. Oxford University Press. ISBN 9780198815419.
- ↑ 10.0 10.1 ">Sweeney, L. (2002). "k-anonymity: A model for protecting privacy". International Journal of Uncertainty, Fuzziness and Knowledge-Based Systems 10 (5): 557–570. doi:10.1142/S0218488502001648.
- ↑ 11.0 11.1 11.2 Liu, F.; Li, T. (2018). "A Clustering K-Anonymity Privacy-Preserving Method for Wearable IoT Devices". Security and Communication Networks 2018: 4945152. doi:10.1155/2018/4945152.
- ↑ 12.0 12.1 12.2 Wei, D.; Ramamurthy, K.N.; Varshney, K.R. (2018). "Distribution‐preserving k‐anonymity". Statistical Analysis and Data Mining 11 (6): 253-270. doi:10.1002/sam.11374.
- ↑ 13.0 13.1 13.2 Lianf, Y.; Samavi, R. (2020). "Optimization-based k-anonymity algorithms". Computers & Security 93: 101753. doi:10.1016/j.cose.2020.101753.
- ↑ 14.0 14.1 14.2 14.3 14.4 Khan, R.; Tao, X.; Anjum, A. et al. (2020). "θ-Sensitive k-Anonymity: An Anonymization Model for IoT based Electronic Health Records". electronics 9 (5): 716. doi:10.3390/electronics9050716.
- ↑ 15.0 15.1 15.2 Li, N.; Li, T.; Venkatasubramanian, S. (2007). "t-Closeness: Privacy Beyond k-Anonymity and l-Diversity". Proceedings of the 2007 IEEE 23rd International Conference on Data Engineering: 106–15. doi:10.1109/ICDE.2007.367856.
- ↑ Nergiz, M.E.; Atzori, M.; Clifton, C. (2007). "Hiding the presence of individuals from shared databases". Proceedings of the 2007 ACM SIGMOD International Conference on Management of Data: 665–76. doi:10.1145/1247480.1247554.
- ↑ Wamg, K.; Fung, B.C.M. (2006). "Anonymizing sequential releases". Proceedings of the 12th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining: 414–23. doi:10.1145/1150402.1150449.
- ↑ Wong, R.C.-W. (2006). "(α, k)-anonymity: An enhanced k-anonymity model for privacy preserving data publishing". Proceedings of the 12th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining: 754–59. doi:10.1145/1150402.1150499.
- ↑ Mohammed, N.; Fung, B.C.M.; Hung, P.C.K. et al. (2009). "Anonymizing healthcare data: A case study on the blood transfusion service". Proceedings of the 15th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining: 1285–94. doi:10.1145/1557019.1557157.
- ↑ Song, F.; Ma, T.; Tian, Y. et al. (2019). "A New Method of Privacy Protection: Random k-Anonymous". IEEE Access 7: 75434-75445. doi:10.1109/ACCESS.2019.2919165.
- ↑ Zhu, H.; Liang, H.-B.; Zhao, L. et al. (2018). "τ-Safe ( l,k )-Diversity Privacy Model for Sequential Publication With High Utility". IEEE Access 7: 687–701. doi:10.1109/ACCESS.2018.2885618.
- ↑ Dwork, C. (2008). "Differential Privacy: A Survey of Results". Proceedings of the 2008 International Conference on Theory and Applications of Models of Computation: 1–19. doi:10.1007/978-3-540-79228-4_1.
- ↑ Alnemari, S.; Romanowski, C.J.; Raj, R.K. (2017). "An Adaptive Differential Privacy Algorithm for Range Queries over Healthcare Data". Proceedings of the 2017 IEEE International Conference on Healthcare Informatics: 397–402. doi:10.1109/ICHI.2017.49.
- ↑ Li, H.; Dai, Y.; Lin, X. (2015). "Efficient e-health data release with consistency guarantee under differential privacy". Proceedings of the 17th International Conference on E-health Networking, Application & Services: 602–608. doi:10.1109/HealthCom.2015.7454576.
- ↑ Guttierrez, O.; Saavedra, J.J.; Zurbaran, M. et al. (2018). "User-Centered Differential Privacy Mechanisms for Electronic Medical Records". Proceedings of the 2018 International Carnahan Conference on Security Technology: 1–5. doi:10.1109/CCST.2018.8585555.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation and grammar for readability. In some cases important information was missing from the references, and that information was added.