Difference between revisions of "Journal:Structure-based knowledge acquisition from electronic lab notebooks for research data provenance documentation"
Shawndouglas (talk | contribs) (Saving and adding more.) |
Shawndouglas (talk | contribs) (Saving and adding more.) |
||
| Line 49: | Line 49: | ||
{{clear}} | {{clear}} | ||
{| | {| | ||
| | | style="vertical-align:top;" | | ||
{| border="0" cellpadding="5" cellspacing="0" width="1200px" | {| border="0" cellpadding="5" cellspacing="0" width="1200px" | ||
|- | |- | ||
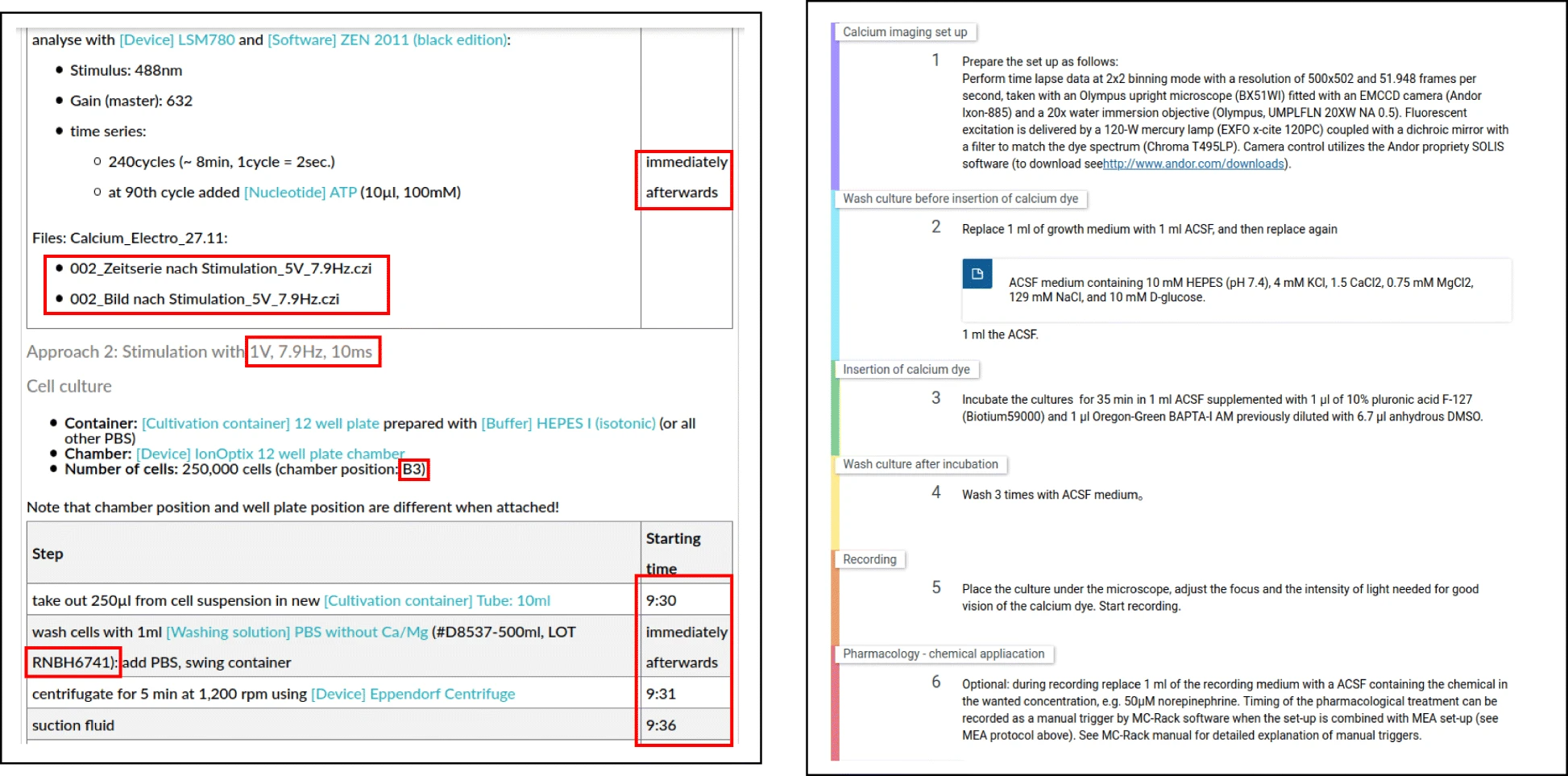
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 1.''' Excerpts of an ELN protocol that represents a particular experiment including all details such as timestamps, lot numbers as well as the research data ('''left''') and a protocol template containing general instructions of experiments without these details ('''right''', sourced [https://doi.org/10.17504/protocols.io.tqfemtn from here].)</blockquote> | | style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Figure 1.''' Excerpts of an ELN protocol that represents a particular experiment including all details such as timestamps, lot numbers as well as the research data ('''left''') and a protocol template containing general instructions of experiments without these details ('''right''', sourced [https://doi.org/10.17504/protocols.io.tqfemtn from here].)</blockquote> | ||
|- | |- | ||
|} | |} | ||
| Line 71: | Line 71: | ||
{{clear}} | {{clear}} | ||
{| | {| | ||
| | | style="vertical-align:top;" | | ||
{| border="0" cellpadding="5" cellspacing="0" width="900px" | {| border="0" cellpadding="5" cellspacing="0" width="900px" | ||
|- | |- | ||
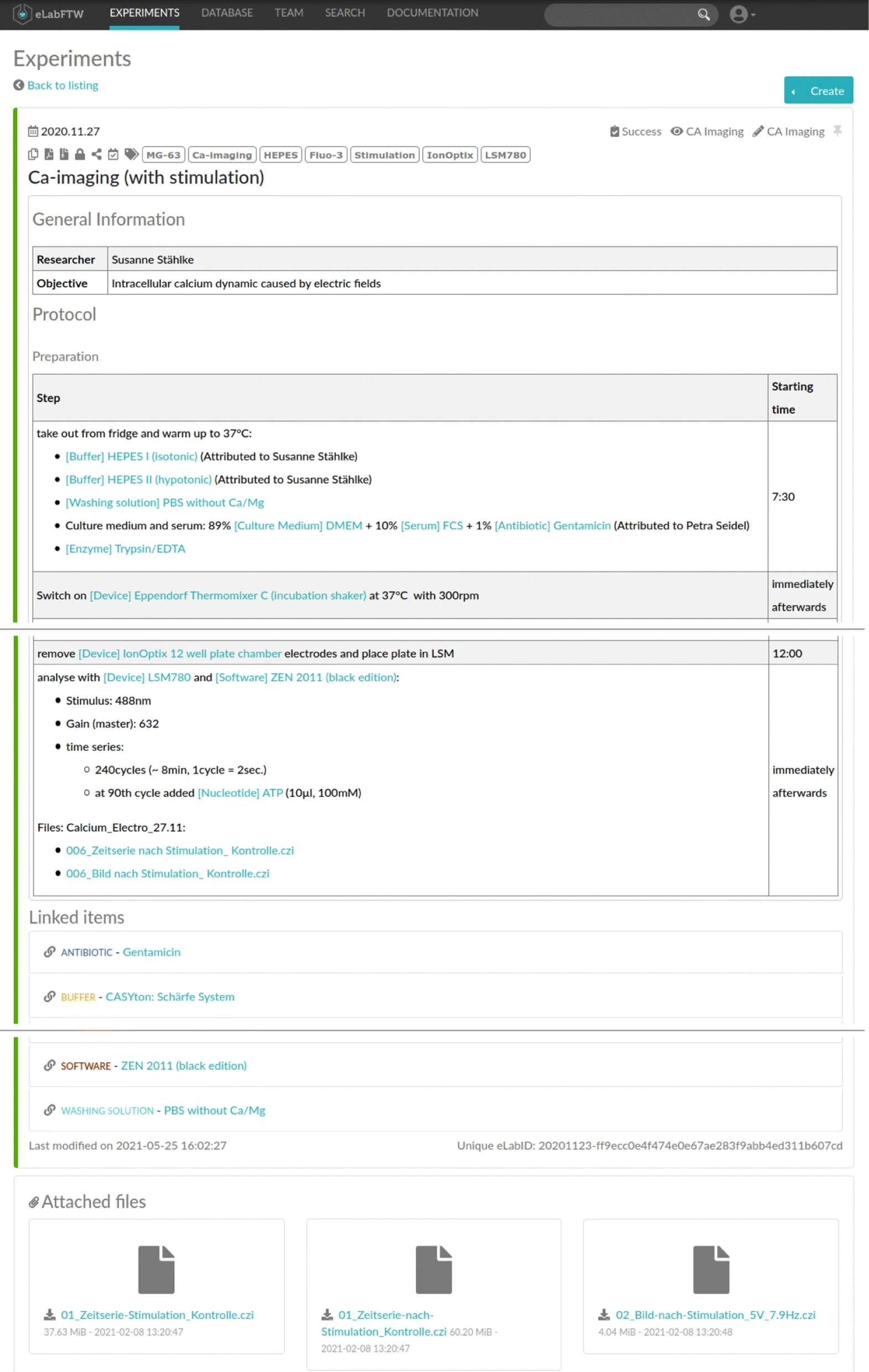
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 2.''' ELN protocol about a Ca-imaging experiment in the eLabFTW software. It contains general information ('''top'''), the list of activities with their starting time ('''middle'''), used inventory items, and uploaded research data ('''bottom''').</blockquote> | | style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Figure 2.''' ELN protocol about a Ca-imaging experiment in the eLabFTW software. It contains general information ('''top'''), the list of activities with their starting time ('''middle'''), used inventory items, and uploaded research data ('''bottom''').</blockquote> | ||
|- | |- | ||
|} | |} | ||
| Line 85: | Line 85: | ||
{{clear}} | {{clear}} | ||
{| | {| | ||
| | | style="vertical-align:top;" | | ||
{| border="0" cellpadding="5" cellspacing="0" width="800px" | {| border="0" cellpadding="5" cellspacing="0" width="800px" | ||
|- | |- | ||
| style="background-color:white; padding-left:10px; padding-right:10px;"| <blockquote>'''Figure 3.''' Shortened documentation of a Ca-imaging experiment in the eLabFTW ELN software. The upper part contains general information about the investigation, followed by the list of activities with their starting time. Below, used inventory items and uploaded research data are listed.</blockquote> | | style="background-color:white; padding-left:10px; padding-right:10px;" |<blockquote>'''Figure 3.''' Shortened documentation of a Ca-imaging experiment in the eLabFTW ELN software. The upper part contains general information about the investigation, followed by the list of activities with their starting time. Below, used inventory items and uploaded research data are listed.</blockquote> | ||
|- | |- | ||
|} | |} | ||
| Line 131: | Line 131: | ||
Many methods aiming at documenting the provenance of activities have already been proposed. Here, we consider the classification of provenance information following the definition of Herschel ''et al.'' [12] and Lim ''et al.'' [13]: | Many methods aiming at documenting the provenance of activities have already been proposed. Here, we consider the classification of provenance information following the definition of Herschel ''et al.'' [12] and Lim ''et al.'' [13]: | ||
# prospective provenance describes “an abstract workflow specification as a recipe for future data derivation” [13]; | #prospective provenance describes “an abstract workflow specification as a recipe for future data derivation” [13]; | ||
# retrospective provenance documents a “past workflow execution and data derivation information, i.e., which tasks were performed and how data artifacts were derived” [13]; and | #retrospective provenance documents a “past workflow execution and data derivation information, i.e., which tasks were performed and how data artifacts were derived” [13]; and | ||
# evolution provenance illustrates “the changes made between two versions of the input” [12], or, in other words, versions of the procedure, the data, or the parameters are reflected by evolution provenance similar to version control such as that implemented by Git for source code. | #evolution provenance illustrates “the changes made between two versions of the input” [12], or, in other words, versions of the procedure, the data, or the parameters are reflected by evolution provenance similar to version control such as that implemented by Git for source code. | ||
Applying those definitions to the use case at hand, prospective provenance allows the keeping track of changes of laboratory-specific operating procedures in general, while retrospective provenance allows the documenting of the actually executed sequence of activities that resulted in a particular set of research data. At last, evolution provenance allows the tracking of changes made to the actual ELN protocol or the inventory database items. | Applying those definitions to the use case at hand, prospective provenance allows the keeping track of changes of laboratory-specific operating procedures in general, while retrospective provenance allows the documenting of the actually executed sequence of activities that resulted in a particular set of research data. At last, evolution provenance allows the tracking of changes made to the actual ELN protocol or the inventory database items. | ||
| Line 139: | Line 139: | ||
With respect to the research workflows to be represented by provenance modeling, two different types can be distinguished: | With respect to the research workflows to be represented by provenance modeling, two different types can be distinguished: | ||
# ''In-silico'' studies employ computational methods for the analysis of the data. [[Workflow]] systems like Taverna [14], Kepler [15], or [[Galaxy (biomedical software)|Galaxy]] [16], and programming environments like [[Jupyter Notebook]] [17] have been successfully augmented to record retrospective provenance. | #''In-silico'' studies employ computational methods for the analysis of the data. [[Workflow]] systems like Taverna [14], Kepler [15], or [[Galaxy (biomedical software)|Galaxy]] [16], and programming environments like [[Jupyter Notebook]] [17] have been successfully augmented to record retrospective provenance. | ||
# Wet lab experiments are courses of activities in a laboratory. While several approaches exist that describe prospective provenance [18, 19] by analyzing published protocols, only limited work is done on documenting retrospective provenance for these workflows. | #Wet lab experiments are courses of activities in a laboratory. While several approaches exist that describe prospective provenance [18, 19] by analyzing published protocols, only limited work is done on documenting retrospective provenance for these workflows. | ||
More detailed information about provenance modelling and the employed methods are provided in the literature. [3, 12] Here, we are interested in providing detailed information about the origin of research data. Thus, we aim at providing retrospective provenance documentation of research data from ELN protocols documenting wet lab experiments. | More detailed information about provenance modelling and the employed methods are provided in the literature. [3, 12] Here, we are interested in providing detailed information about the origin of research data. Thus, we aim at providing retrospective provenance documentation of research data from ELN protocols documenting wet lab experiments. | ||
The Smart Tea project [20] similarly aims at the semantic metadata recording for research data from within a customized ELN. The developed ELN provides a structured graphical user interface (GUI) requiring the user to provide information for predefined variables. All information is directly transferred into a linked data representation and persistently archived with a linked data server. While this approach perfectly guides users through the sequence of activities and tracks retrospective provenance at the same time, it fails to keep track of deviations from the predefined plan. Furthermore, as the documentation is directly translated into a semantic representation, additional information that was not considered before can hardly be attached to such protocols, which restricts both the expressivity of the semantic model and the user to previously known information. | The Smart Tea project [20] similarly aims at the semantic metadata recording for research data from within a customized ELN. The developed ELN provides a structured graphical user interface (GUI) requiring the user to provide information for predefined variables. All information is directly transferred into a linked data representation and persistently archived with a linked data server. While this approach perfectly guides users through the sequence of activities and tracks retrospective provenance at the same time, it fails to keep track of deviations from the predefined plan. Furthermore, as the documentation is directly translated into a semantic representation, additional information that was not considered before can hardly be attached to such protocols, which restricts both the expressivity of the semantic model and the user to previously known information. | ||
Similar to the Smart Tea project, the PROV templating approach [21] suggests the recording of provenance information given a pre-defined provenance model. In other words, the main idea is that applications only store values for placeholders in a particular provenance model, which was shown to be more efficient than the storage of the original provenance models. [21] This solution is very efficient if a very large number of identical provenance structures with some variable information are to be stored. If, however, the application requires more flexibility in terms of the provenance structure, the template approach does not utilize this efficiency advantage. Note that provenance templates encode a semantic representation with variables, whereas protocol templates provide guidelines for experiments. | |||
Curcin ''et al.'' [22] use a very similar approach for the provenance modelling in [[Clinical decision support system|diagnostic decision support systems]]. A more flexible approach is the use of knowledge graph cells (KGCs), proposed by Vogt ''et al.'' [23] They provide a concept for the definition of knowledge structures. In particular, rules including ABox and TBox expressions might be defined that allow the dynamic modification of the KG. Thus, KGCs might be used to specify potential semantic structures of ELN protocols without particular information inside. The application of KGCs would require a complete definition over all possible semantic representations of ELN protocols, which is infeasible. | |||
With respect to the vocabulary used to semantically describe the laboratory-specific information, the EXperimental ACTions (EXACT2) ontology, together with the Natural Language Processing (NLP) framework [18], aims at the automatic extraction of knowledge from biomedical protocols for prospective provenance. Similarly, the SeMAntic RepresenTation for Experimental Protocols (SMART Protocols) ontology reuses EXACT2 to represent prospective provenance from published protocols. [19] In contrast to both approaches that represent a plan, we aim at retrospective provenance, i.e., a particular course of activities. Both approaches, however, could be used to describe prospective provenance of the underlying plan of an ELN protocol, to allow the documentation of potential deviations from the original plan. The Reproduce Microscopy Experiments (REPRODUCE-ME) ontology [24] introduces a specific vocabulary to describe retrospective provenance for microscopy experiments. Besides, the domain-independent ontologies, PROV-O and its predecessor Open Provenance Model (OPM) [25], are frequently employed as upper-level ontology for provenance documentation. [3] Furthermore, many extensions for specific applications have been proposed. The Provenance, Authoring, and Versioning (PAV) ontology, for example, proposes a mechanism for the versioning and authoring of web resources [26], and CollabPG encodes collaborations within processes. [3] With respect to the application domain of the use case, the Open Biological and Biomedical Ontology (OBO) Foundry is a community initiative aiming at the development and maintenance of ontologies in the biomedical domain. [27] The Basic Formal Ontology (BFO) [28] is the upper-level ontology that is used for each of the OBO ontologies. | |||
For the retrospective provenance documentation of research data from computational workflows, several specifically tailored tools and approaches have been proposed in the literature. ProvBook [17], for instance, tracks provenance in Jupyter notebooks that are used for literate programming. There's also Dataprov [29], a wrapper tool producing provenance information from the execution of analysis tools, and noWorkflow [30], which captures provenance information from analysis scripts such as for the programming language Python. Aside from these methods, other provenance tracking approaches known as lineage retrieval [31] or lineage tracking and workflow systems exist. [32] In general, ''in-silico'' workflow systems not only record provenance information, but at the same time they specify the involved processing steps and enable their execution possibly on a distributed system. [33] However, as these systems are limited to tackling computational analyses, their usage for the provenance of research data from wet lab experiments is difficult. | |||
Regarding the completeness of the documentation with respect to reproducibility, plenty of standards exist that aim at the definition of the minimum set of information required to comprehend and reproduce the research investigation for different applications. With respect to the use case at hand, the minimum information for electrical cell stimulation [34] and the Minimum Information About a Cellular Assay (MIACA)<ref name="MIACA">{{cite web |url=http://miaca.sourceforge.net/ |title=MIACA - Minimum Information About a Cellular Assay |author=MIACA Standards Initiative |work=SourceForge |date=2006}}</ref> provide such references for the documentation. Similarly, standard operating procedures (SOPs) or published instructions for experiments encode standards for the documentation of a particular experiment. | |||
When considering the publication or archiving of research data, metadata is important to provide additional context, enabling others (including the future self) to understand the research process and the resulting data. In particular, the FAIR guiding principles provide abstract recommendations for handling research data to enable its re-usability. [1] Together with the implementation suggestions of these guidelines [2], they provide a framework which is also applicable for research data from wet lab experiments. While both guidelines provide generic recommendations regarding research data documentation, different standards exist that provide vocabulary for their support. Several initiatives foster the development of documentation standards for research data, including the Data Documentation Initiative (DDI) that focuses on standardizing metadata for social science datasets. [35] The Dublin Core, instead, is a more general definition of 15 metadata elements for electronic resources. [36, 37] Similarly, Data Catalog Vocabulary (DCAT) provides a common vocabulary for the interoperability of data catalogs [38] and, thus, also defines required metadata for research data. Additionally, domain-specific metadata standards have been developed. With respect to the use case, this includes metadata for microscopy images, such as that proposed by the RDM4mic Initiative.<ref>{{Cite journal |last=Kunis, S. |date=22 October 2021 |title=Workgroup RDM4mic - Research data management for microscopy |url=https://zenodo.org/record/5591958 |journal=Zenodo |doi=10.5281/zenodo.5591958}}</ref> In addition to these metadata, the information inside the data file might also be described. For this purpose, codebooks and data dictionaries are employed. [39, 40] Considering a CSV file as an example, this includes information about each column such as the domain of the values and the unit of the measurements. This information is defined in a separate file that helps comprehend the raw data. | |||
For the publication and archiving of this data, including the semantic documentation, several approaches have been proposed. These include bundling formats such as BagIt [41], Oxford Common File Layout (OCFL) [42], and RO-Crate [43], as well as literate programming methods such as using Jupyter Notebook to combine (parts of) research data, their analysis source code, and results, as well as their documentation. RO-Crate [43] is a mechanism that allows the bundling of resources together with their associated metadata, supporting the FAIR publication and archiving of the research data. By re-using existing vocabulary such as schema.org or PROV-O, it implements a linked data approach to enable researchers to provide all information necessary to (re-)use the described research data. This includes basic properties such as author and title of the resource, a license for publication, a description of the files, and a description of the workflow used to create those files in terms of retrospective provenance, including employed software and other equipment. In brief, a RO-Crate bundle consists of the research data file and a metadata file called <tt>ro-crate-metadata.json</tt>, which contains structured metadata about the files and the entire bundle in a JSON-LD format. While the <tt>ro-crate-metadata.json</tt> contains all information in machine interpretable way, it is accompanied by a human readable HTML representation. RO-Crate has successfully been used for the documentation of retrospective provenance of ''in-silico'' studies [44], but can, due to the flexibility of the vocabulary, also be used for retrospective provenance of wet lab experiments. | |||
==Methods== | |||
Revision as of 22:54, 30 March 2022
| Full article title | Structure-based knowledge acquisition from electronic lab notebooks for research data provenance documentation |
|---|---|
| Journal | Journal of Biomedical Semantics |
| Author(s) | Schröder, Max; Staehlke, Susanne; Groth, Paul; Nebe, J. Barbara; Spors, Sascha; Krüger, Frank |
| Author affiliation(s) | University of Rostock, University Medical Center Rostock, University of Amsterdam |
| Primary contact | Email: max dot schroeder at uni-rostock dot de |
| Year published | 2022 |
| Volume and issue | 13 |
| Article # | 4 (2022) |
| DOI | 10.1186/s13326-021-00257-x |
| ISSN | 2041-1480 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://jbiomedsem.biomedcentral.com/articles/10.1186/s13326-021-00257-x |
| Download | https://jbiomedsem.biomedcentral.com/track/pdf/10.1186/s13326-021-00257-x.pdf (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
Background: Electronic laboratory notebooks (ELNs) are used to document experiments and investigations in the wet lab. Protocols in ELNs contain a detailed description of the conducted steps, including the necessary information to understand the procedure and the raised research data, as well as to reproduce the research investigation. The purpose of this study is to investigate whether such ELN protocols can be used to create semantic documentation of the provenance of research data by the use of ontologies and linked data methodologies.
Methods: Based on an ELN protocol of a biomedical wet lab experiment, a retrospective provenance model of the raised research data describing the details of the experiment in a machine-interpretable way is manually engineered. Furthermore, an automated approach for knowledge acquisition from ELN protocols is derived from these results. This structure-based approach exploits the structure in the experiment’s description—such as headings, tables, and links—to translate the ELN protocol into a semantic knowledge representation. To satisfy the FAIR guiding principles (making data findable, accessible, interoperable, and reuseable), a ready-to-publish bundle is created that contains the research data together with their semantic documentation.
Results: While the manual modelling efforts serve as proof of concept by employing one protocol, the automated structure-based approach demonstrates the potential generalization with seven ELN protocols. For each of those protocols, a ready-to-publish bundle is created and, by employing the SPARQL query language, it is illustrated such that questions about the processes and the obtained research data can be answered.
Conclusions: The semantic documentation of research data obtained from the ELN protocols allows for the representation of the retrospective provenance of research data in a machine-interpretable way. Research Object Crate (RO-Crate) bundles including these models enable researchers to easily share the research data, including the corresponding documentation, as well as to search and relate the experiment to each other.
Keywords: research data, provenance, knowledge acquisition, electronic laboratory notebooks, semantic documentation, RO-Crate, FAIR
Background
Effective reuse of research data requires comprehensive documentation of their provenance. Beside metadata, knowledge about the generating process helps others to understand research data and allows for the reproduction of research investigations. This includes not only sources of input data, such as parameters and assumptions, but also information about instrumentation, devices, and materials. For wet lab experiments, such knowledge is increasingly documented in electronic laboratory notebooks (ELNs). The focus of these tools is on the documentation of laboratory activities that produce research data in so-called "ELN protocols." In addition to this textual description, the FAIR guiding principles [1] provide general guidance on research data documentation in terms of metadata. However, they do not prescribe technical details about the implementation of such documentation. [2]
To foster the realization of the FAIR principles for research data produced in wet lab experiments, we aim for machine-interpretable representations of experimental documentation of the process that is the origin of the data. In other words, the provenance information about the research data—including the activities and involved researchers, resources, and equipment—should be semantically represented. For this purpose, we employ the frequently used [3] PROV W3C recommendation [4], which ontologically, in PROV Ontology (PROV-O), defines entities, activities, and agents, as well as their relations. In particular, according to Belhajjame et al., an entity is defined as a “physical, digital, conceptual, or other kind of thing with some fixed aspects,” [5] an activity as “something that occurs over a period of time and acts upon or with entities; it may include consuming, processing, transforming, modifying, relocating, using, or generating entities,” [5] and an agent as “something that bears some form of responsibility for an activity taking place, for the existence of an entity, or for another agent’s activity.” [5] With respect to wet lab experiments, all biological and chemical resources—as well as not only the devices and software but also the research data itself—can be seen as entities; researchers conducting the experiment are the agents, and the process of research data creation consists of activities. The semantic representation of this information as a knowledge graph (KG) [6] can be achieved by the use of modern web technologies where the terms and their relations are defined in ontologies such as PROV-O (TBox modelling), the instances are built up in the KG (ABox modelling), and other KGs can be linked in order to create an interconnected graph of semantic knowledge.
In this paper, we aim for an automatic extraction of information from ELN protocols in order to transfer them into a semantic representation that documents the produced research data. For this purpose, we employ the documentation of Calcium imaging (Ca-imaging) experiments, originally proposed by Staehlke et al. [7], as a running example. In particular, we use ELN protocols that document the conduction of Ca-imaging experiments in order to: (i) demonstrate the feasibility of manually transferring an ELN protocol into a semantic representation encoding the provenance of research data, (ii) automate the information extraction and modelling by exploiting the structure of an ELN protocol by means of a structure-based approach, and (iii) evaluate the proposed method by answering provenance questions from the resulting bundle of research data and the corresponding semantic model.
Here, the term "ELN protocol" refers to the actual documentation of the wet lab experiment within an ELN and is different from the term "protocol templates," which are used to encode instructions to be performed in order to conduct particular procedures or be published at protocols.io. While those protocol templates do encode a list of abstract instructions, they do not necessarily reflect particular research data, nor instrumentation, parameters, or other aspects to the execution-specific information. ELN protocols, in contrast, represent the documentation of the actual experiment, and the contained information is thus necessary to understand how the resulting research data was generated. This includes manufacturer-specific information about resources used in the experiment such as lot numbers.[a] Furthermore, passage numbers of the resources, the times when an activity was conducted, and the parameters used in a device, as well as the research data and the researchers conducting the experiment, are information specific to a particular experiment. Figure 1 illustrates the differences by providing an example for an ELN protocol and a protocol template.
|
The work presented here is based on a preliminary investigation regarding the effectiveness of manually modeling ELN protocols by use of ontologies. [8] Here, we extend this preliminary work by discussing the potential of automatic information extraction from ELN protocols by employing structural information and discussing the differences and implications of both approaches. Moreover, while the previous work only sketched the semantic representation of the wet lab experiments, here, we focus on the generation of ready-to-publish research data bundles, including the semantic description of the origin of the research data.
Use case
To demonstrate the feasibility of the proposed approach, a typical wet lab investigation was chosen as a use case. In the following, we introduce the use case and derive questions regarding the provenance of the corresponding research data.
Biomedical wet lab experiments
The objective of the biomedical study was to investigate the intracellular calcium ions (Ca2+) dynamics by Calcium-imaging (Ca-imaging) under different settings. [7] In particular, two different wet lab experiments were considered: (i) an investigation of the influence of different material surface conditions on Ca2+ mobilization, and (ii) an investigation regarding the Ca2+ dynamics under the influence of electrical stimulation. Both types of experiments involve similar activities of the researchers. In particular, each experiment employs the Ca-imaging method previously established by Staehlke et al. [7] in different settings. The particular conditions, e.g., surface conditions or parameters of the electrical stimulation, are investigated within each experiment, while the order of the different variations was permuted across the experiments. That is, after a preparation phase, where all materials and devices are prepared, the same procedure, i.e., Ca-imaging, was executed for the different conditions. During the experiment, several materials and devices are employed, such as cell line passages, buffer, and microscopes.
For the purpose of this study, we asked the researchers to use an ELN for the documentation of their wet lab activities, resulting in eight ELN protocols: one for the first experiment and seven for the latter, representing different permutations of the sequential execution of Ca-imaging for different electrical stimulation parameters. In particular, eLabFTW (Deltablot, https://www.elabftw.net/, v3.6.7) [10], a domain-independent ELN, was used. Figure 2 shows an excerpt of a protocol from the use case.
|
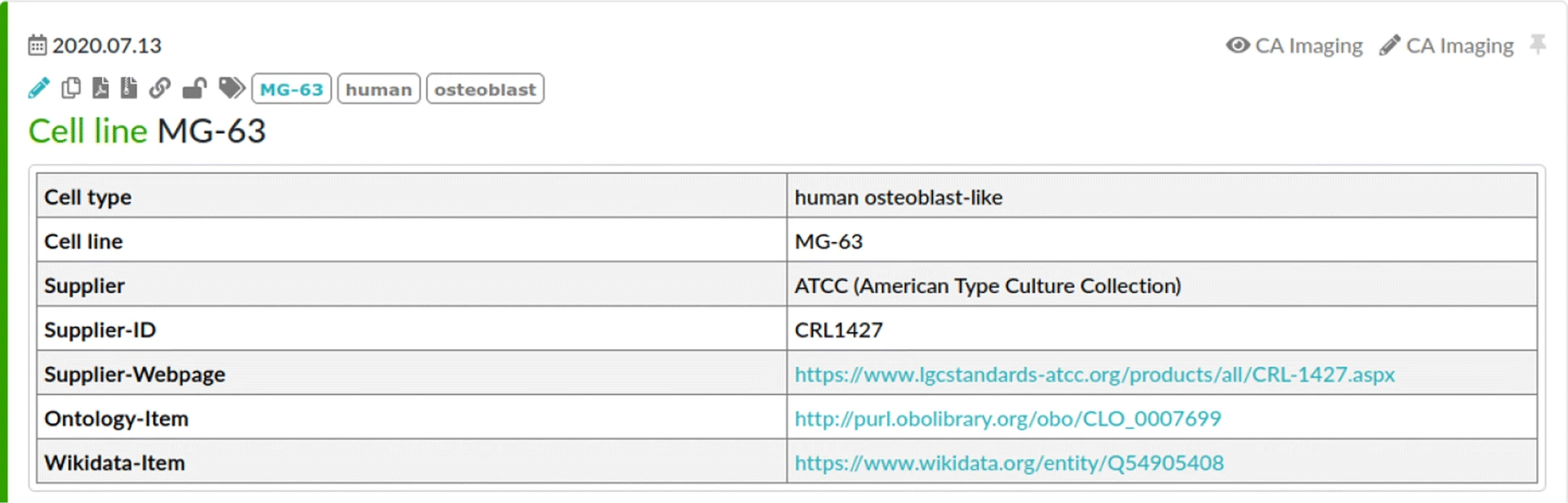
ELNs often provide an inventory database that allows the maintenance of materials and other research resources used during the experiments. Typically, each resource belongs to a configurable set of categories, e.g., cell lines, buffer, software, or devices. These entries in the inventory database can be linked from within the protocol when used within the corresponding experiment. Figure 3 illustrates the entry to the inventory database for the MG-63 cell line that is used in the experiments of the use case. Note that this entry is already augmented by information about ontology classes that were added during the manual model engineering process. Here, we use such ontology references but also other resource identifiers, such as Research Resource Identifiers (RRID)[1], could be used for resource reference. However, these RRIDs do not reflect different versions of the resources, e.g., when describing a software. Thus, they can be used to annotate the inventory database of the ELN similar to the ontology classes, but cannot be used on their own. Research data is attached to the ELN protocol by uploading and linking from within the textual description of the step that describes the generating activity.
|
In summary, the execution of an individual experiment took about 4.5 hours, resulting from the preparation and the sequential executions of the Ca-imaging procedure under five different stimulation settings consisting of 15 steps for each. Each protocol referred to 22 inventory items in the database and between 85 and 110 data files of different types were generated. The different file types include (i) CZI files (developed by ZEISS) containing the microscope settings, recorded images, and raw measurement data; (ii) image files in JPEG format to illustrate particular excerpts from the video recordings; and (iii) raw measurements of the luminescence over time, in the form of XML encoded tabular data files. The latter two formats are exports from the CZI files. The provenance of all attached files needs to be documented.
Research data provenance
When considering this use case, several questions regarding the provenance of the research data can be raised. To this end, we consider questions based on the W7 provenance model [11], that describes provenance as combinations of What, When, Where, How, Who, Which, and Why. We consider each question individually, encoding the view of a researcher that aims at re-using the research data from our use case. The questions were developed together with the domain experts and resemble actual questions that arise when considering the replication of the documented experiments.
- W1 Who participated in the study?
- With respect to the provenance of research data, all researchers contributing to the creation are of interest, i.e., we expect to get a list of all researchers and their affiliations involved in an experiment.
- W2 Which biological and chemical resources and which equipment was used in the study?
- In particular, we are interested in the resources and the equipment used in an experiment, including all details such as the lot number and the passage information.
- W3 How was a particular file created?
- "What was the sequence of activities that led to the creation of a particular file" is a question that might help other researchers in comprehending the data.
- W4 When was an activity conducted?
- The date and the time point of a particular activity but also its duration are of interest. This information is useful for the planning of similar experiments, but also with respect to the comprehensibility of the results as the date and time point might influence them, e.g., due to weather or other environmental phenomena.
- W5 Why was the experiment done?
- Understanding why the research data was created is crucial for their comprehensibility. We take the objective of the experiment as the reason for the creation.
- W6 Where was the experiment conducted?
- The location—respectively. the institution where the experiment was conducted—is of interest as regional characteristics might influence the data.
- W7 What was the order of the stimulation parameters in a particular experiment?
- The order of the particular approaches influences the results as there might be effects from the timing of the experiments or the duration since their preparation. That means, with respect to the evaluation of the results, we are interested in this order.
Related work
The provenance of research data, including their research investigations, combines several research fields, ranging from general-purpose methods and standards for the documentation of provenance to specifically tailored methods and platforms for the tracking of research and other activities. In the following, we will discuss recent work within those fields and relate it to our method.
Many methods aiming at documenting the provenance of activities have already been proposed. Here, we consider the classification of provenance information following the definition of Herschel et al. [12] and Lim et al. [13]:
- prospective provenance describes “an abstract workflow specification as a recipe for future data derivation” [13];
- retrospective provenance documents a “past workflow execution and data derivation information, i.e., which tasks were performed and how data artifacts were derived” [13]; and
- evolution provenance illustrates “the changes made between two versions of the input” [12], or, in other words, versions of the procedure, the data, or the parameters are reflected by evolution provenance similar to version control such as that implemented by Git for source code.
Applying those definitions to the use case at hand, prospective provenance allows the keeping track of changes of laboratory-specific operating procedures in general, while retrospective provenance allows the documenting of the actually executed sequence of activities that resulted in a particular set of research data. At last, evolution provenance allows the tracking of changes made to the actual ELN protocol or the inventory database items.
With respect to the research workflows to be represented by provenance modeling, two different types can be distinguished:
- In-silico studies employ computational methods for the analysis of the data. Workflow systems like Taverna [14], Kepler [15], or Galaxy [16], and programming environments like Jupyter Notebook [17] have been successfully augmented to record retrospective provenance.
- Wet lab experiments are courses of activities in a laboratory. While several approaches exist that describe prospective provenance [18, 19] by analyzing published protocols, only limited work is done on documenting retrospective provenance for these workflows.
More detailed information about provenance modelling and the employed methods are provided in the literature. [3, 12] Here, we are interested in providing detailed information about the origin of research data. Thus, we aim at providing retrospective provenance documentation of research data from ELN protocols documenting wet lab experiments.
The Smart Tea project [20] similarly aims at the semantic metadata recording for research data from within a customized ELN. The developed ELN provides a structured graphical user interface (GUI) requiring the user to provide information for predefined variables. All information is directly transferred into a linked data representation and persistently archived with a linked data server. While this approach perfectly guides users through the sequence of activities and tracks retrospective provenance at the same time, it fails to keep track of deviations from the predefined plan. Furthermore, as the documentation is directly translated into a semantic representation, additional information that was not considered before can hardly be attached to such protocols, which restricts both the expressivity of the semantic model and the user to previously known information.
Similar to the Smart Tea project, the PROV templating approach [21] suggests the recording of provenance information given a pre-defined provenance model. In other words, the main idea is that applications only store values for placeholders in a particular provenance model, which was shown to be more efficient than the storage of the original provenance models. [21] This solution is very efficient if a very large number of identical provenance structures with some variable information are to be stored. If, however, the application requires more flexibility in terms of the provenance structure, the template approach does not utilize this efficiency advantage. Note that provenance templates encode a semantic representation with variables, whereas protocol templates provide guidelines for experiments.
Curcin et al. [22] use a very similar approach for the provenance modelling in diagnostic decision support systems. A more flexible approach is the use of knowledge graph cells (KGCs), proposed by Vogt et al. [23] They provide a concept for the definition of knowledge structures. In particular, rules including ABox and TBox expressions might be defined that allow the dynamic modification of the KG. Thus, KGCs might be used to specify potential semantic structures of ELN protocols without particular information inside. The application of KGCs would require a complete definition over all possible semantic representations of ELN protocols, which is infeasible.
With respect to the vocabulary used to semantically describe the laboratory-specific information, the EXperimental ACTions (EXACT2) ontology, together with the Natural Language Processing (NLP) framework [18], aims at the automatic extraction of knowledge from biomedical protocols for prospective provenance. Similarly, the SeMAntic RepresenTation for Experimental Protocols (SMART Protocols) ontology reuses EXACT2 to represent prospective provenance from published protocols. [19] In contrast to both approaches that represent a plan, we aim at retrospective provenance, i.e., a particular course of activities. Both approaches, however, could be used to describe prospective provenance of the underlying plan of an ELN protocol, to allow the documentation of potential deviations from the original plan. The Reproduce Microscopy Experiments (REPRODUCE-ME) ontology [24] introduces a specific vocabulary to describe retrospective provenance for microscopy experiments. Besides, the domain-independent ontologies, PROV-O and its predecessor Open Provenance Model (OPM) [25], are frequently employed as upper-level ontology for provenance documentation. [3] Furthermore, many extensions for specific applications have been proposed. The Provenance, Authoring, and Versioning (PAV) ontology, for example, proposes a mechanism for the versioning and authoring of web resources [26], and CollabPG encodes collaborations within processes. [3] With respect to the application domain of the use case, the Open Biological and Biomedical Ontology (OBO) Foundry is a community initiative aiming at the development and maintenance of ontologies in the biomedical domain. [27] The Basic Formal Ontology (BFO) [28] is the upper-level ontology that is used for each of the OBO ontologies.
For the retrospective provenance documentation of research data from computational workflows, several specifically tailored tools and approaches have been proposed in the literature. ProvBook [17], for instance, tracks provenance in Jupyter notebooks that are used for literate programming. There's also Dataprov [29], a wrapper tool producing provenance information from the execution of analysis tools, and noWorkflow [30], which captures provenance information from analysis scripts such as for the programming language Python. Aside from these methods, other provenance tracking approaches known as lineage retrieval [31] or lineage tracking and workflow systems exist. [32] In general, in-silico workflow systems not only record provenance information, but at the same time they specify the involved processing steps and enable their execution possibly on a distributed system. [33] However, as these systems are limited to tackling computational analyses, their usage for the provenance of research data from wet lab experiments is difficult.
Regarding the completeness of the documentation with respect to reproducibility, plenty of standards exist that aim at the definition of the minimum set of information required to comprehend and reproduce the research investigation for different applications. With respect to the use case at hand, the minimum information for electrical cell stimulation [34] and the Minimum Information About a Cellular Assay (MIACA)[2] provide such references for the documentation. Similarly, standard operating procedures (SOPs) or published instructions for experiments encode standards for the documentation of a particular experiment.
When considering the publication or archiving of research data, metadata is important to provide additional context, enabling others (including the future self) to understand the research process and the resulting data. In particular, the FAIR guiding principles provide abstract recommendations for handling research data to enable its re-usability. [1] Together with the implementation suggestions of these guidelines [2], they provide a framework which is also applicable for research data from wet lab experiments. While both guidelines provide generic recommendations regarding research data documentation, different standards exist that provide vocabulary for their support. Several initiatives foster the development of documentation standards for research data, including the Data Documentation Initiative (DDI) that focuses on standardizing metadata for social science datasets. [35] The Dublin Core, instead, is a more general definition of 15 metadata elements for electronic resources. [36, 37] Similarly, Data Catalog Vocabulary (DCAT) provides a common vocabulary for the interoperability of data catalogs [38] and, thus, also defines required metadata for research data. Additionally, domain-specific metadata standards have been developed. With respect to the use case, this includes metadata for microscopy images, such as that proposed by the RDM4mic Initiative.[3] In addition to these metadata, the information inside the data file might also be described. For this purpose, codebooks and data dictionaries are employed. [39, 40] Considering a CSV file as an example, this includes information about each column such as the domain of the values and the unit of the measurements. This information is defined in a separate file that helps comprehend the raw data.
For the publication and archiving of this data, including the semantic documentation, several approaches have been proposed. These include bundling formats such as BagIt [41], Oxford Common File Layout (OCFL) [42], and RO-Crate [43], as well as literate programming methods such as using Jupyter Notebook to combine (parts of) research data, their analysis source code, and results, as well as their documentation. RO-Crate [43] is a mechanism that allows the bundling of resources together with their associated metadata, supporting the FAIR publication and archiving of the research data. By re-using existing vocabulary such as schema.org or PROV-O, it implements a linked data approach to enable researchers to provide all information necessary to (re-)use the described research data. This includes basic properties such as author and title of the resource, a license for publication, a description of the files, and a description of the workflow used to create those files in terms of retrospective provenance, including employed software and other equipment. In brief, a RO-Crate bundle consists of the research data file and a metadata file called ro-crate-metadata.json, which contains structured metadata about the files and the entire bundle in a JSON-LD format. While the ro-crate-metadata.json contains all information in machine interpretable way, it is accompanied by a human readable HTML representation. RO-Crate has successfully been used for the documentation of retrospective provenance of in-silico studies [44], but can, due to the flexibility of the vocabulary, also be used for retrospective provenance of wet lab experiments.
Methods
Footnotes
- ↑ A lot number is an identifier for a particular set of materials produced by one manufacturer. Thus, lot numbers enable to track information about the provenance of these material productions.
References
- ↑ "RRID Portal". SciCrunch. 2021. https://scicrunch.org/resources.
- ↑ MIACA Standards Initiative (2006). "MIACA - Minimum Information About a Cellular Assay". SourceForge. http://miaca.sourceforge.net/.
- ↑ Kunis, S. (22 October 2021). "Workgroup RDM4mic - Research data management for microscopy". Zenodo. doi:10.5281/zenodo.5591958. https://zenodo.org/record/5591958.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. To more easily differentiate footnotes from references, the original footnotes (which were numbered) were updated to use lowercase letters. Most footnotes referencing web pages were turned into proper citations.