Journal:Thirty years of the DICOM standard
| Full article title | Thirty years of the DICOM standard |
|---|---|
| Journal | Tomography |
| Author(s) | Larobina, Michele |
| Author affiliation(s) | Consiglio Nazionale delle Ricerche |
| Primary contact | Email: michele dot larobina at cnr dot it |
| Year published | 2023 |
| Volume and issue | 9(5) |
| Page(s) | 1829-1838 |
| DOI | 10.3390/tomography9050145 |
| ISSN | 2379-139X |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.mdpi.com/2379-139X/9/5/145 |
| Download | https://www.mdpi.com/2379-139X/9/5/145/pdf?version=1696571652 (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
Digital Imaging and Communications in Medicine (DICOM) is an international standard that defines a format for storing medical images and a protocol to enable and facilitate data communication among medical imaging systems. The DICOM standard has been instrumental in transforming the medical imaging world over the last three decades. Its adoption has been a significant experience for manufacturers, healthcare users, and research scientists. In this review, 30 years after introducing the standard, we discuss the innovation, advantages, and limitations of adopting DICOM and its possible future directions.
Keywords: DICOM, metadata, file formats, communication protocols, quantitative imaging
Introduction
Standardization is a key concept in the digital imaging world. The absence of a standard limits the usability and sharing of medical images. It forces users to deal with a multitude of data formats and to convert data from one format to another. Moreover, any image file, in addition to pixel data, contains metadata. Metadata is data describing the image and plays a non-secondary role in digital imaging. While general-purpose image format metadata can be limited to the description of the pixel matrix, in formats for scientific applications metadata can describe the subject, the instrumentation set-up, the image acquisition parameters, and any other element of interest related to the imaging workflow. Despite this, the power of metadata is most often underestimated and consequently unexpressed. A standard helps in defining the metadata section for the correct use and interpretation of the image itself.
The field of medical imaging is exemplary in the context of standardization processes for pioneering vision and for having created a long-lived and appreciated standard. In the early 1980s, an association of users and professionals of the healthcare sector, the American College of Radiology (ACR), jointly with the National Association of Electronic Manufacturers (NEMA), started to define a new standard for the encoding and exchange of digital medical images. In 1993, the ACR-NEMA committee presented Digital Imaging and Communications in Medicine (DICOM) as a standard with more functionality and long-term vision than the previous standardization attempts known as ACR-NEMA 1.0 (1985) and 2.0 (1988). [1,2,3,4,5,6,7] For its time, DICOM represented an authentic novelty. Before the introduction of the DICOM standard, and therefore, until the first half of the 1990s, the medical imaging world saw diagnostic modalities, even within the same department, very confined to their rooms. The images were generally printed to film to be interpreted by the radiologist. In a native digital format, images were viewed and processed on the modality console and rarely exported to different workstations. Medical imaging systems were not connected to each other except for some dedicated point-to-point connections. Image transfer between modalities and image processing workstation, both inside and outside an imaging department, took place mainly through removable media with an important limitation: the absence of a common file format and the unknown of correctly reading the removable media storage (typically a magneto-optical disks or a tape). Therefore, the usability of the images remained linked to the availability of software for reading proprietary image formats and the media themselves. (A complete overview of the old proprietary file formats can be found on the David Clunie Medical Image Formats website. [8]) The DICOM international standard was conceived to overcome the limitations imposed by proprietary architectures and data formats and to allow and promote communication among medical imaging devices using the same network infrastructures of internet-enabled computer networks.
With its conception, the novelties of the DICOM standard were:
- To state that in medicine the standardization of image format and image-related information, as well as their communication over a network, is essential;
- To remark that, for medical images, metadata is as important as pixel data; and
- To be general enough to cover almost every medical imaging modality and flexible enough to follow their evolution over time.
First developed for radiology and then cardiology departments, the DICOM standard has evolved over the years to support various other branches of medical imaging well beyond radiology, such as dermatology and ophthalmology, with the objective to encompass almost all modalities of imaging-based medicine. There are approximately 80 modalities defined by the standard today. DICOM is also the current standard for radiation therapy in the so-called second-generation radiotherapy after a complete revision in 2014. DICOM also supports data exchange of time-based signals or waveforms, such as those generated in clinical neurophysiology, which include, among others, electrocardiograms (ECGs) and electroencephalography (EEG). In more recent times, the DICOM standard has been proposed for digital pathology. The adoption of the standard in this area would favor the integration of clinical imaging and laboratory medicine. [9,10,11] DICOM does not limit its action to images and associated information originating directly from medical devices. It defines mechanisms for archiving and sharing quantitative derived images, image-derived data, annotations, and reports.
DICOM is a constantly evolving standard and is revised five times a year with contributions from the numerous DICOM working groups divided by fields of application and imaging modalities [15]. However, DICOM only provides recommendations and no accompanying software. Despite this, the availability of some high-quality open-source software libraries and utilities in several programming languages—such as DCMTK (C and C++), DCM4CHE (Java), and PyDICOM (Python) [12,13,14]—has helped in spreading and affirming the standard.
This article presents an overview of the DICOM standard, covering its fundamental principles and concepts. It also explores the advantages and limitations of the standard while outlining some potential future developments. Throughout this document, all references to the DICOM standard in sections, figures, and tables refer to the 2023b edition.
Not only pixels: The power of metadata in medical images
Medical images must be standardized in a format that can be stored, shared, and used effectively. Therefore, a standard must necessarily deal with metadata as well. The DICOM standard aims to establish a reliable format for medical images and associated information. One of the most significant advancements of the DICOM image format is its metadata formulation, which provides an accurate and detailed description of the subject and procedure used to generate the image. The standard emphasizes that metadata is essential for the full use of medical investigation for clinical, management, and research purposes, establishing the non-divisibility of the pixel data from the metadata. Each DICOM image consists of metadata and pixel data embedded in a single file so that, as the standard institutional website remarks, “the image can never be separated from this information by mistake.”
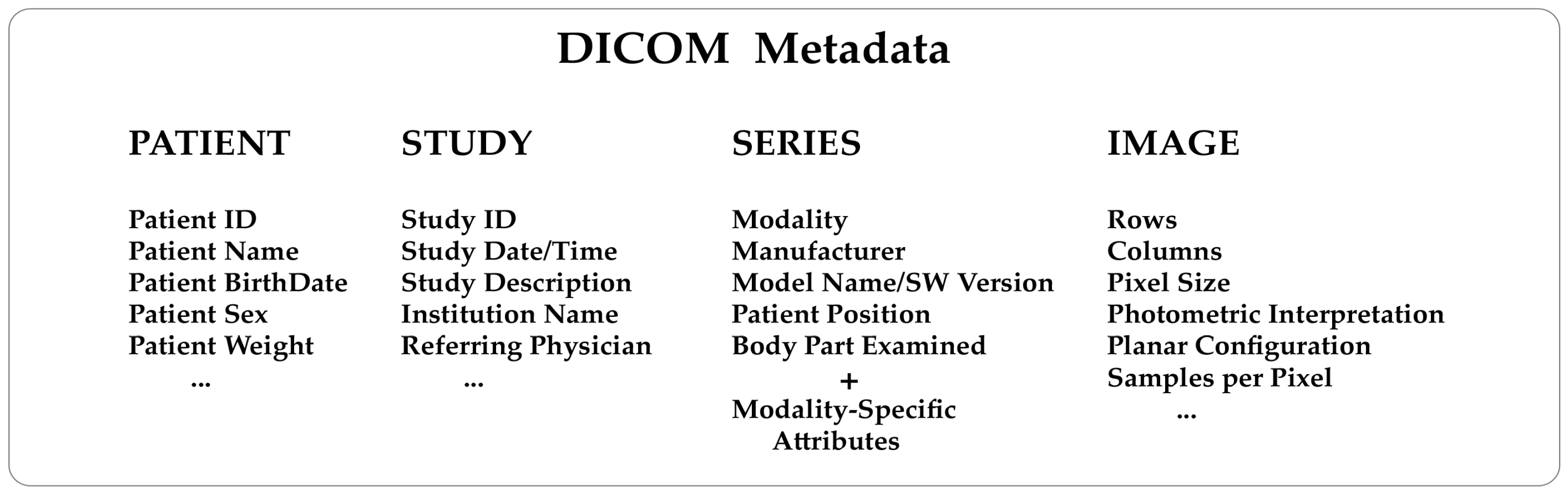
Knowing how a medical image has been generated from the diagnostic modalities is extremely helpful. For example, a nuclear medicine image will contain information about the injected radiopharmaceutical, injection time, acquisition start time, and end time. An X-ray computed tomography image will contain information about the X-ray tube voltage and current, exposure time, slice thickness, etc. This kind of data are consistently included in the metadata of every DICOM image. To understand the structure of a DICOM metadata section, remember that the standard describes real-world entities, such as patients, studies, diagnostic modalities, and images, in terms of objects and relationships that may occur between them (the so-called entity-relationship model). It is a high level of abstraction that has helped make the DICOM a complex format. Objects are defined in a standard way through groups of attributes describing them in detail. As a result, DICOM needs to store many attributes (Figure 1) to guarantee a comprehensive depiction of the imaging process.
|
How is the metadata section populated? There are attributes set during the installation/configuration of the imaging modality (mainly related to the hardware and software of the system, the institution name, etc.), attributes automatically exchanged with the hospital information system (HIS)/radiology information system (RIS) of the department, attributes specified by the acquisition procedure selected for the examination, etc.
In a DICOM file, each attribute is identified by a unique tag consisting of two hexadecimal numbers. The first represents the group and the second represents the element. For example, the tag for the modality is (0008, 0060).
DICOM attributes are logically grouped into modules to identify and describe real-world objects. Modules are presented in standard documents in the form of tables. Modules store the name, tag, definition, and type of the attributes and can hold attributes from various groups. There are modules common to multiple imaging modalities and others specific to one modality, as outlined in Annex C.7 and C.8 of the PS 3.3 standard document (Table 1). Groups of attributes repeated across multiple modules are called "macros." The DICOM data dictionary (PS 3.6) contains all the attributes defined and described in the standard, known as public data elements. Metadata can also include manufacturer-specific attributes known as "private data elements" for which there is no description and are not part of the data dictionary. It is important to note that private data elements are assigned an odd group number, whereas public data elements always have an even group number. The number of data elements present in a DICOM file, both public and private, is generally variable. A specific tag indicates the start of the pixel data and hence the end of the metadata section.
| ||||||
Regarding the DICOM pixel data section, the support for floating point values (single precision 32-bit and double precision 64-bit) is limited to radiation dose values in radiotherapy and, more recently, to parametric maps defined as images in which the pixel values have been derived from the value stored by the modality to be the expression of a physical quantity. In all the other cases, DICOM pixel values can only be integers. DICOM uses a scale factor whenever the values stored in each voxel need to be scaled to different units. This is achieved through two fields specified in the metadata defining the slope and the intercept of the linear transformation to be used to convert pixel values to real-world values.
Beyond a standardized file format for medical images and associated information, the DICOM standard provides a communication protocol to easily share images in a vendor-independent manner. Protocols are defined by Tim Berners-Lee, one of the creators of the World Wide Web, as simple rules for global systems. [16] This definition is both brief and impactful.
The purpose of the DICOM protocol is to establish communication between diagnostic and sometimes therapy systems of different manufacturers and display, storage, and management devices on a network. The introduction of the DICOM standard marks the beginning of a revolution similar to the introduction of computer networks: no more separate diagnostic equipment, but diagnostic systems, and in some cases of therapy, processing/display and reporting stations that can be connected together and that can share images and related data, storage devices and printers. To realize this objective, the DICOM standard provides an upper-layer protocol that runs over the well-established internet standard protocol TCP/IP. Any DICOM-compliant device attached to the network is an Application Entity identified, in addition to the TCP/IP parameters (IP address, subnet mask, and port number) by a 16-character identification code called “Application Entity Title.” Application Entities can exchange services among themselves according to the client-server model that DICOM renames "Service/Provider"; the requestor of a service is called a "Service Class User," while the provider is called a "Service Class Provider." Depending on the service, a DICOM node can act as a user or provider. Establishing an association involves a negotiation phase during which the service to be exchanged and the role played by each node is established. Next, the transfer syntax for the data exchange is decided, the connection is established, and the data transfer takes place. The main services available on a DICOM network are:
- Storage: This service is required to archive images across a network. Typically, it is used by an acquisition modality to send images to a picture archiving and communication system (PACS) or a storage server.
- Storage Commitment: This is an enhanced version of the Storage service with in addition a message sent by the storage provider to the user to confirm that “archiving was successful,” so that the user can safely delete the images locally.
- Print: This service prints images from an acquisition modality or a display station.
- Query/Retrieve: This service enables nodes on the DICOM network to query a PACS or another storage unit in order to know the list of images available on it and then to retrieve them.
- Modality Worklist: This service manages the list of exams to be acquired for each patient. Each examination of the list is scheduled and its completion is made known to the system that updates the data. It is only possible in departments equipped with a computerized reservation/acceptance system (HIS/RIS) integrated into the DICOM network.
Recently, DICOM has added a protocol called DICOMweb built on top of the HyperText Transfer Protocol (HTTP) for using services via the web. [17,18,19,20] DICOMweb enables query, retrieval, storage, and worklist services. DICOM images can be retrieved traditionally as binary objects containing metadata and pixel data or with metadata in JavaScript Object Notation (JSON) or Extensible Markup Language (XML) and pixel data as DICOM bulk data or, optionally, in a format suitable to be directly displayed in a web browser (JPEG).
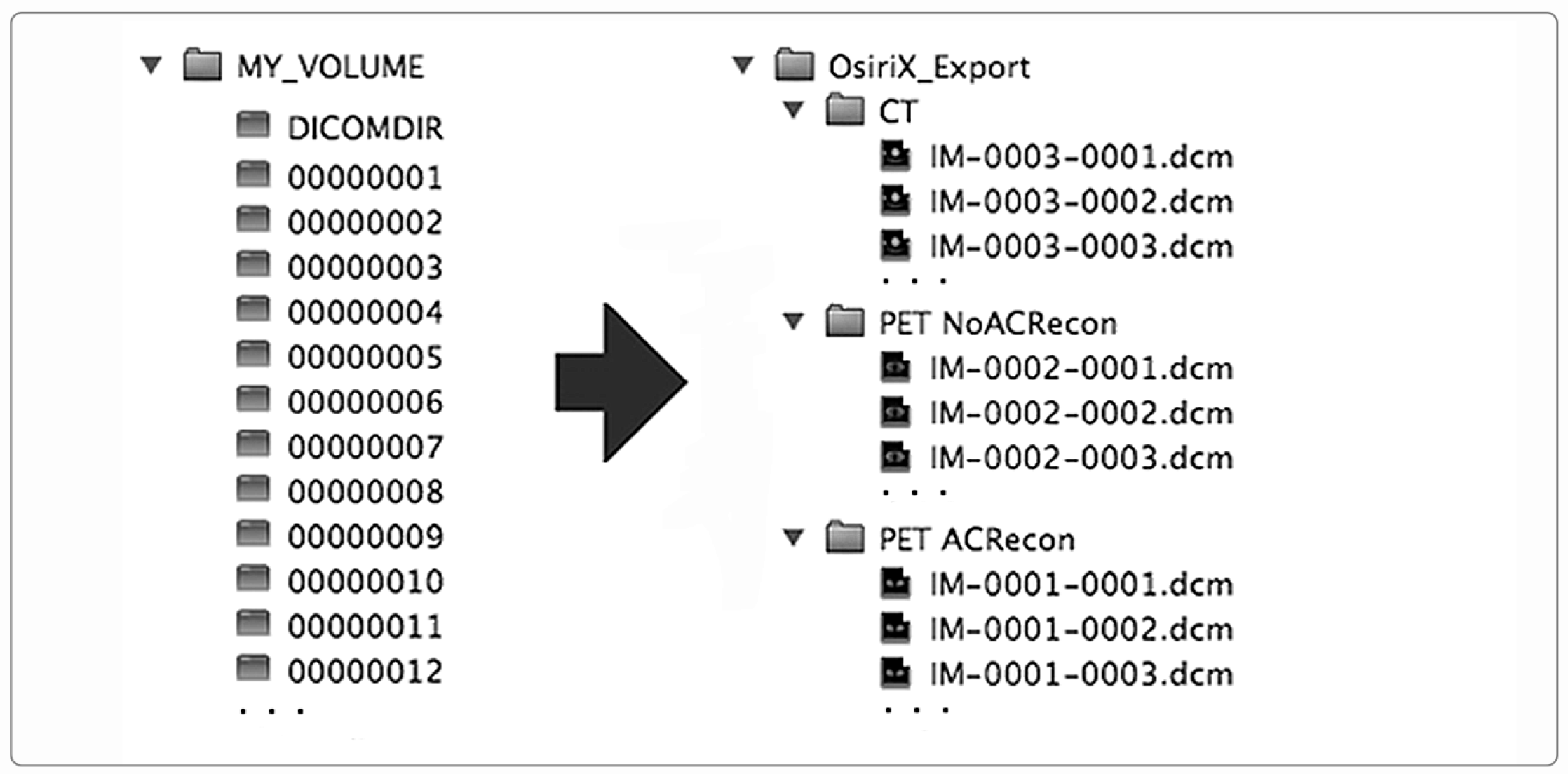
In addition to the storage of images through a network, the DICOM standard specifies how to standardize image storage on removable physical media such as CD/DVD/Blue-ray discs, magneto-optical discs, USB-connected removable devices, and compact-flash removable devices (DICOM PS 3.12). In this way, the interoperability is extended and ensured even when the image exchange takes place through removable supports. To store DICOM images on physical supports, the standard prescribes a flat-file organization consisting of a folder that serves as a container for the patient’s images, along with a DICOMDIR file. DICOMDIR contains the association between image files and the patient study-series information for all the DICOM files on the media. DICOMDIR is not human-readable as it is a binary file in DICOM format, so a software utility is necessary to read it. Figure 2 shows the DICOMDIR concept schematically.
|
DICOM’s strengths and weaknesses
References
Notes
This presentation is faithful to the original, with only a few minor changes to presentation, spelling, and grammar. In some cases important information was missing from the references, and that information was added. The original put information into a Table 2 that didn't need to be in a table; it was made into a bulleted list for this version.