Featured article of the week: March 28–April 03:
"The need for informatics to support forensic pathology and death investigation"
As a result of their practice of medicine, forensic pathologists create a wealth of data regarding the causes of and reasons for sudden, unexpected or violent deaths. This data have been effectively used to protect the health and safety of the general public in a variety of ways despite current and historical limitations. These limitations include the lack of data standards between the thousands of death investigation (DI) systems in the United States, rudimentary electronic information systems for DI, and the lack of effective communications and interfaces between these systems. Collaboration between forensic pathology and clinical informatics is required to address these shortcomings and a path forward has been proposed that will enable forensic pathology to maximize its effectiveness by providing timely and actionable information to public health and public safety agencies. (Full article...)
Featured article of the week: March 21–27:
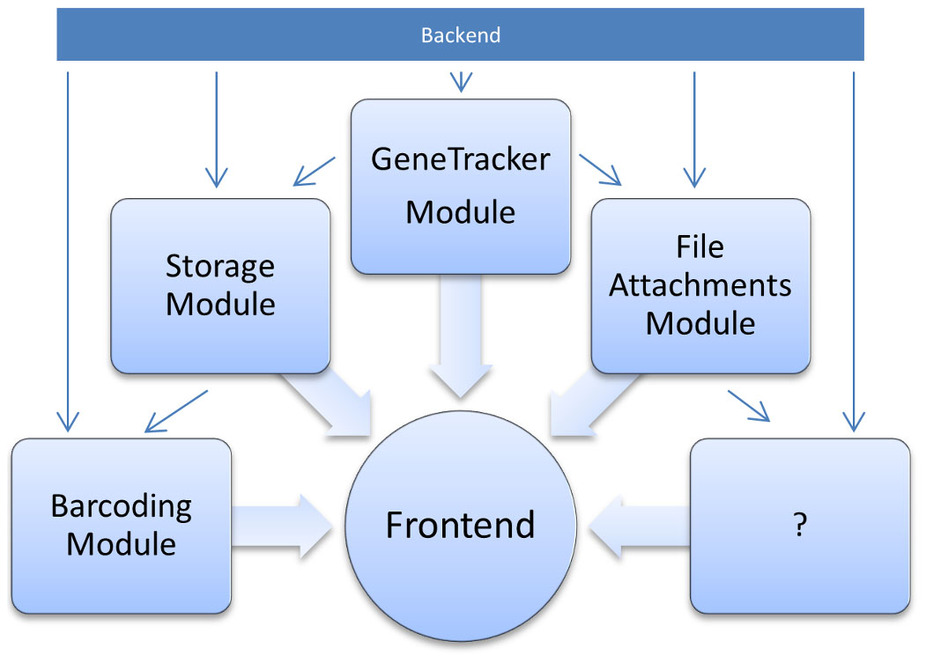
"Efficient sample tracking with OpenLabFramework"
The advance of new technologies in biomedical research has led to a dramatic growth in experimental throughput. Projects therefore steadily grow in size and involve a larger number of researchers. Spreadsheets traditionally used are thus no longer suitable for keeping track of the vast amounts of samples created and need to be replaced with state-of-the-art laboratory information management systems. Such systems have been developed in large numbers, but they are often limited to specific research domains and types of data. One domain so far neglected is the management of libraries of vector clones and genetically engineered cell lines. OpenLabFramework is a newly developed web-application for sample tracking, particularly laid out to fill this gap, but with an open architecture allowing it to be extended for other biological materials and functional data. Its sample tracking mechanism is fully customizable and aids productivity further through support for mobile devices and barcoded labels. (Full article...)
|
Featured article of the week: March 14–20:
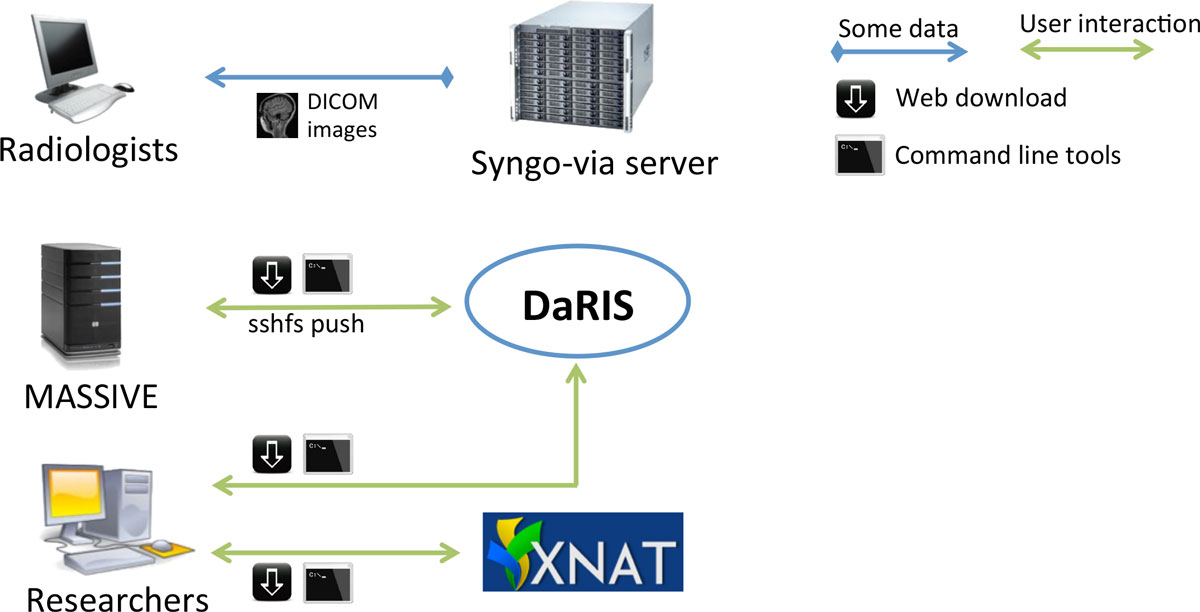
"Design, implementation and operation of a multimodality research imaging informatics repository"
Biomedical imaging research increasingly involves acquiring, managing and processing large amounts of distributed imaging data. Integrated systems that combine data, meta-data and workflows are crucial for realising the opportunities presented by advances in imaging facilities. This paper describes the design, implementation and operation of a multi-modality research imaging data management system that manages imaging data obtained from biomedical imaging scanners operated at Monash Biomedical Imaging (MBI), Monash University in Melbourne, Australia. In addition to Digital Imaging and Communications in Medicine (DICOM) images, raw data and non-DICOM biomedical data can be archived and distributed by the system. Imaging data are annotated with meta-data according to a study-centric data model and, therefore, scientific users can find, download and process data easily. The research imaging data management system ensures long-term usability, integrity inter-operability and integration of large imaging data. Research users can securely browse and download stored images and data, and upload processed data via subject-oriented informatics frameworks including the Distributed and Reflective Informatics System (DaRIS), and the Extensible Neuroimaging Archive Toolkit (XNAT). (Full article...)
|
Featured article of the week: March 07–13:
Forensic science (often shortened to forensics) is the application of a broad spectrum of sciences — from anthropology to toxicology — to answer questions of interest to a legal system. During the course of an investigation, forensic scientists collect, preserve, and analyze scientific evidence using a variety of special laboratory equipment and special techniques for such interests. In addition to their laboratory role, the forensic scientists may also testify as an expert witness in both criminal and civil cases and can work for either the prosecution or the defense.
Much of the work of forensic science is conducted in the forensic laboratory. Such a laboratory has many similarities to a traditional clinical or research lab in so much that it contains various lab instruments and several areas set aside for different tasks. However, it differs in other ways. Windows, for example, represent a point of entry into a forensic lab, which must be secure as it contains evidence to crimes. (Full article...)
|
Featured article of the week: February 29–March 06:
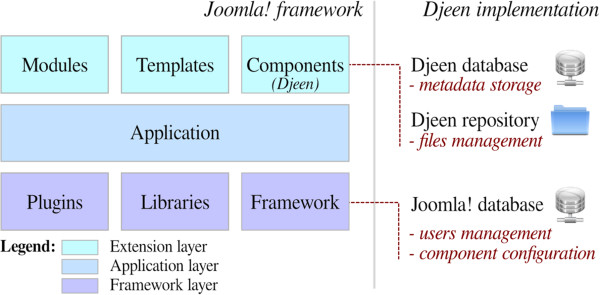
"Djeen (Database for Joomla!’s Extensible Engine): A research information management system for flexible multi-technology project administration"
With the advance of post-genomic technologies, the need for tools to manage large scale data in biology becomes more pressing. This involves annotating and storing data securely, as well as granting permissions flexibly with several technologies (all array types, flow cytometry, proteomics) for collaborative work and data sharing. This task is not easily achieved with most systems available today.
We developed Djeen (Database for Joomla!’s Extensible Engine), a new Research Information Management System (RIMS) for collaborative projects. Djeen is a user-friendly application, designed to streamline data storage and annotation collaboratively. Its database model, kept simple, is compliant with most technologies and allows storing and managing of heterogeneous data with the same system. Advanced permissions are managed through different roles. Templates allow Minimum Information (MI) compliance. (Full article...)
|
Featured article of the week: February 22–28:
"SeqWare Query Engine: Storing and searching sequence data in the cloud"
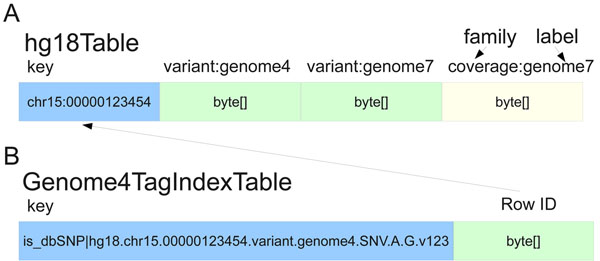
Since the introduction of next-generation DNA sequencers the rapid increase in sequencer throughput, and associated drop in costs, has resulted in more than a dozen human genomes being resequenced over the last few years. These efforts are merely a prelude for a future in which genome resequencing will be commonplace for both biomedical research and clinical applications. The dramatic increase in sequencer output strains all facets of computational infrastructure, especially databases and query interfaces. The advent of cloud computing, and a variety of powerful tools designed to process petascale datasets, provide a compelling solution to these ever increasing demands.
In this work, we present the SeqWare Query Engine which has been created using modern cloud computing technologies and designed to support databasing information from thousands of genomes. Our backend implementation was built using the highly scalable, NoSQL HBase database from the Hadoop project. We also created a web-based frontend that provides both a programmatic and interactive query interface and integrates with widely used genome browsers and tools. (Full article...)
|
Featured article of the week: February 15–21:
"SaDA: From sampling to data analysis—An extensible open source infrastructure for rapid, robust and automated management and analysis of modern ecological high-throughput microarray data"
One of the most crucial characteristics of day-to-day laboratory information management is the collection, storage and retrieval of information about research subjects and environmental or biomedical samples. An efficient link between sample data and experimental results is absolutely important for the successful outcome of a collaborative project. Currently available software solutions are largely limited to large scale, expensive commercial Laboratory Information Management Systems (LIMS). Acquiring such LIMS indeed can bring laboratory information management to a higher level, but most of the times this requires a sufficient investment of money, time and technical efforts. There is a clear need for a light weighted open source system which can easily be managed on local servers and handled by individual researchers. Here we present a software named SaDA for storing, retrieving and analyzing data originated from microorganism monitoring experiments. SaDA is fully integrated in the management of environmental samples, oligonucleotide sequences, microarray data and the subsequent downstream analysis procedures. It is simple and generic software, and can be extended and customized for various environmental and biomedical studies. (Full article...)
|
Featured article of the week: January 25–31:
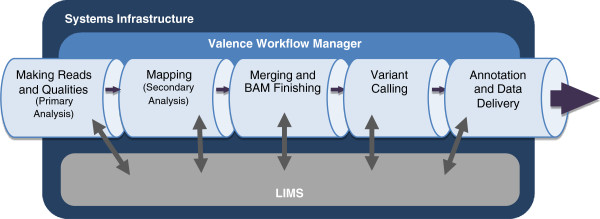
"Launching genomics into the cloud: Deployment of Mercury, a next generation sequence analysis pipeline"
Massively parallel DNA sequencing generates staggering amounts of data. Decreasing cost, increasing throughput, and improved annotation have expanded the diversity of genomics applications in research and clinical practice. This expanding scale creates analytical challenges: accommodating peak compute demand, coordinating secure access for multiple analysts, and sharing validated tools and results.
To address these challenges, we have developed the Mercury analysis pipeline and deployed it in local hardware and the Amazon Web Services cloud via the DNAnexus platform. Mercury is an automated, flexible, and extensible analysis workflow that provides accurate and reproducible genomic results at scales ranging from individuals to large cohorts.
By taking advantage of cloud computing and with Mercury implemented on the DNAnexus platform, we have demonstrated a powerful combination of a robust and fully validated software pipeline and a scalable computational resource that, to date, we have applied to more than 10,000 whole genome and whole exome samples. (Full article...)
|
Featured article of the week: January 18–24:
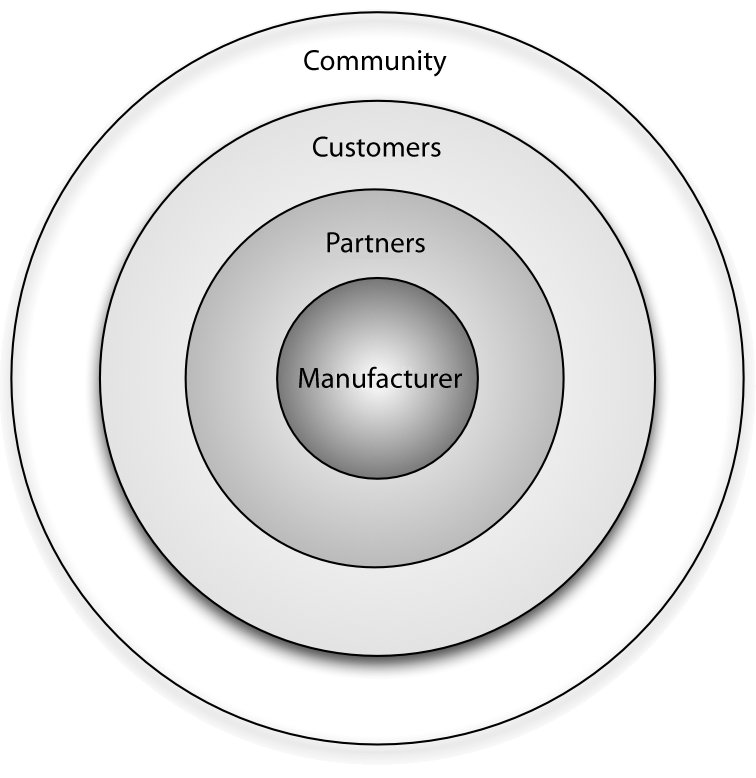
"Benefits of the community for partners of open source vendors"
Open source vendors can benefit from business ecosystems that form around their products. Partners of such vendors can utilize this ecosystem for their own business benefit by understanding the structure of the ecosystem, the key actors and their relationships, and the main levers of profitability. This article provides information on all of these aspects and identifies common business scenarios for partners of open source vendors. Armed with this information, partners can select a strategy that allows them to participate in the ecosystem while also maximizing their gains and driving adoption of their product or solution in the marketplace.
Every free/libre open source software (F/LOSS) vendor strives to create a business ecosystem around its software product. Doing this offers two primary advantages from a sales and marketing perspective: i) it increases the viability and longevity of the product in both commercial and communal spaces, and ii) it opens up new channels for communication and innovation. (Full article...)
|
Featured article of the week: January 11–17:
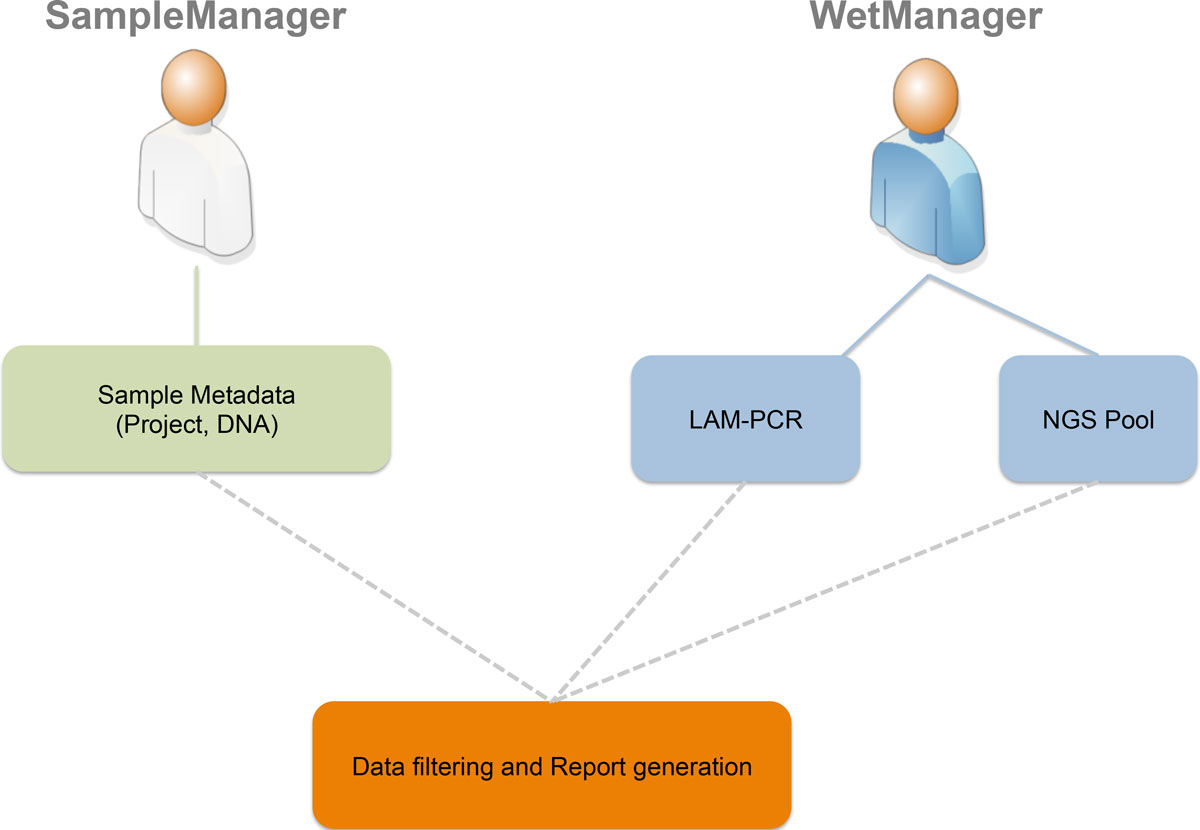
"adLIMS: A customized open source software that allows bridging clinical and basic molecular research studies"
Many biological laboratories that deal with genomic samples are facing the problem of sample tracking, both for pure laboratory management and for efficiency. Our laboratory exploits PCR techniques and Next Generation Sequencing (NGS) methods to perform high-throughput integration site monitoring in different clinical trials and scientific projects. Because of the huge amount of samples that we process every year, which result in hundreds of millions of sequencing reads, we need to standardize data management and tracking systems, building up a scalable and flexible structure with web-based interfaces, which are usually called Laboratory Information Management System (LIMS).
We extended and customized ADempiere ERP to fulfill LIMS requirements and we developed adLIMS. It has been validated by our end-users verifying functionalities and GUIs through test cases for PCRs samples and pre-sequencing data and it is currently in use in our laboratories. adLIMS implements authorization and authentication policies, allowing multiple users management and roles definition that enables specific permissions, operations and data views to each user. (Full article...)
|
Featured article of the week: January 4–10:
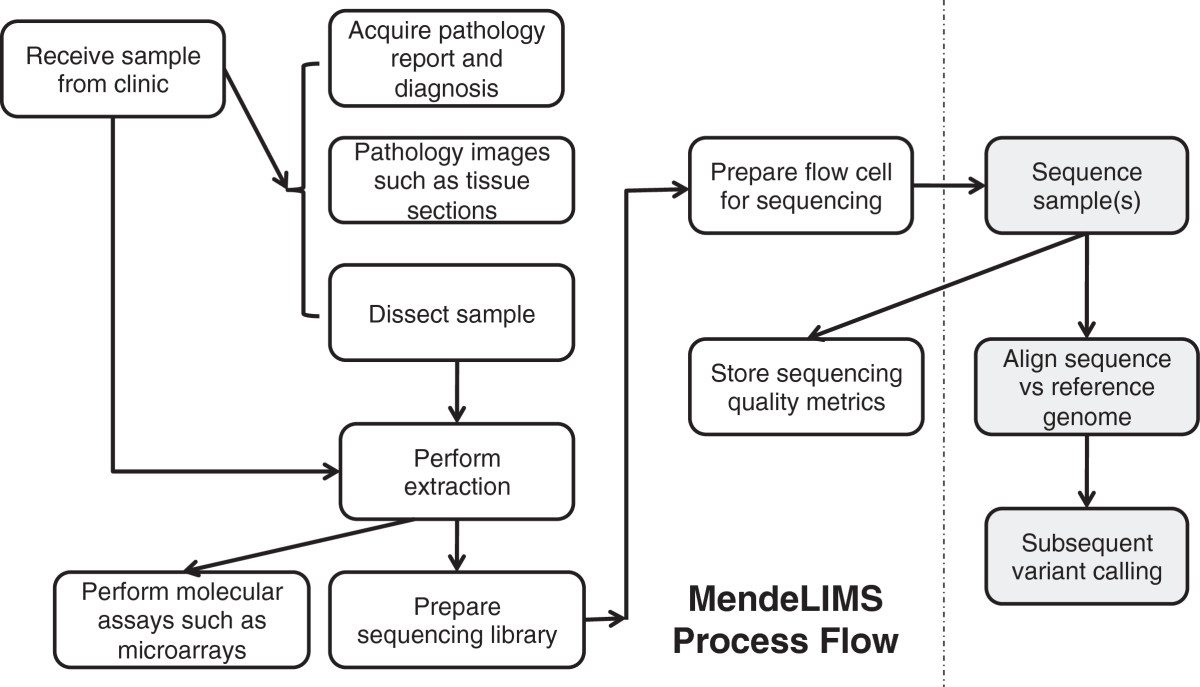
"MendeLIMS: A web-based laboratory information management system for clinical genome sequencing"
Large clinical genomics studies using next generation DNA sequencing require the ability to select and track samples from a large population of patients through many experimental steps. With the number of clinical genome sequencing studies increasing, it is critical to maintain adequate laboratory information management systems to manage the thousands of patient samples that are subject to this type of genetic analysis.
To meet the needs of clinical population studies using genome sequencing, we developed a web-based laboratory information management system (LIMS) with a flexible configuration that is adaptable to continuously evolving experimental protocols of next generation DNA sequencing technologies. Our system is referred to as MendeLIMS, is easily implemented with open source tools and is also highly configurable and extensible. MendeLIMS has been invaluable in the management of our clinical genome sequencing studies. (Full article...)
|
|