Journal:Bridging data management platforms and visualization tools to enable ad-hoc and smart analytics in life sciences
| Full article title | Bridging data management platforms and visualization tools to enable ad-hoc and smart analytics in life sciences |
|---|---|
| Journal | Journal of Integrative Bioinformatics |
| Author(s) | Panse, Christian; Trachsel, Christian; Türker, Can |
| Author affiliation(s) | Functional Genomics Center Zurich |
| Primary contact | Email: cp at fgcz dot ethz dot ch |
| Year published | 2022 |
| Volume and issue | 19(4) |
| Article # | 20220031 |
| DOI | 10.1515/jib-2022-0031 |
| ISSN | 1613-4516 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.degruyter.com/document/doi/10.1515/jib-2022-0031/html |
| Download | https://www.degruyter.com/document/doi/10.1515/jib-2022-0031/pdf (PDF) |
|
|
This article contains rendered mathematical formulae. You may require the TeX All the Things plugin for Chrome or the Native MathML add-on and fonts for Firefox if they don't render properly for you. |
Abstract
Core facilities, which share centralized research resources across institutions and organizations, have to offer technologies that best serve the needs of their users and provide them a competitive advantage in research. They have to set up and maintain tens to hundreds of instruments, which produce large amounts of data and serve thousands of active projects and customers. Particular emphasis has to be given to the reproducibility of the results. Increasingly, the entire process—from building the research hypothesis, conducting the experiments, and taking the measurements, through to data exploration and analysis—is solely driven by very few experts in various scientific fields. Still, the ability to perform data exploration entirely in real-time on a personal computer is often hampered by the heterogeneity of software, data structure formats of the output, and the enormous data file sizes. These impact the design and architecture of the implemented software stack.
At the Functional Genomics Center Zurich (FGCZ), a joint state-of-the-art research and training facility of ETH Zurich and the University of Zurich, we have developed the B-Fabric system, which has served for more than a decade an entire life sciences community with fundamental data science support. In this paper, we describe how such a system can be used to glue together data (including metadata), computing infrastructures (clusters and clouds), and visualization software to support instant data exploration and visual analysis. We illustrate our implemented daily approach using visualization applications of mass spectrometry (MS) data.
Keywords: accessible, findable, interoperable, and reusable (FAIR); integrations for data analysis; open research data (ORD); workflow
Introduction
Core facilities—which act as a discrete, centralized location for shared research resources for institutions or organizations[1]—aim to support scientific research where terabytes of archivable raw data are routinely produced every year. This data is annotated, pre-processed[2], quality-controlled[3], and analyzed, ideally within research pipelines. Additionally, reports are generated and charges are sent to the customers to conclude the entire support process. Typically, data acquisition, management, and analysis cycles are conducted in parallel for multiple projects with different research questions, involving several experts and scientists from different fields, such as biology, medicine, chemistry, statistics, and computer science, as well as various fields of omics.
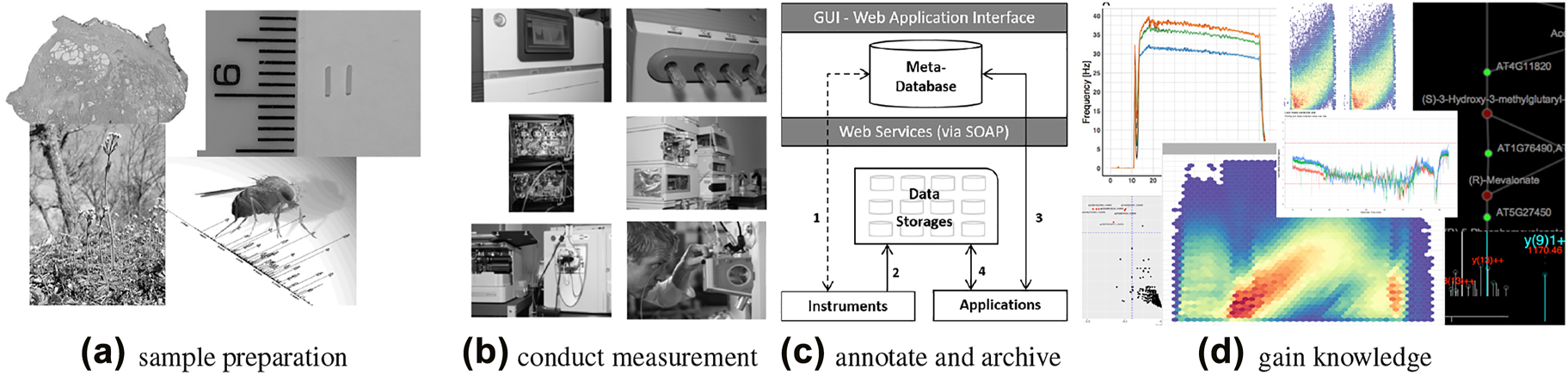
Figure 1 sketches a typical omics workflow: (a) Samples of different organisms and tissues (e.g., a fresh frozen human tissue sample with indicated cancer and benign prostate tissue areas, or an Arabidopsis or Drosophila melanogaster[4] model organism) are collected. (b) Sequencing and mass spectrometry (MS) are typical methods of choice to perform measurement based on the prepared samples in the omics fields. (c) Data handling and archiving occurs, including metadata, with special emphasis on the reproducibility of the measurement results. (d) Largely known community-developed bioinformatics tools and visualization techniques are applied ad-hoc to gain knowledge, exploiting not only the raw data but also the associated metadata.
|
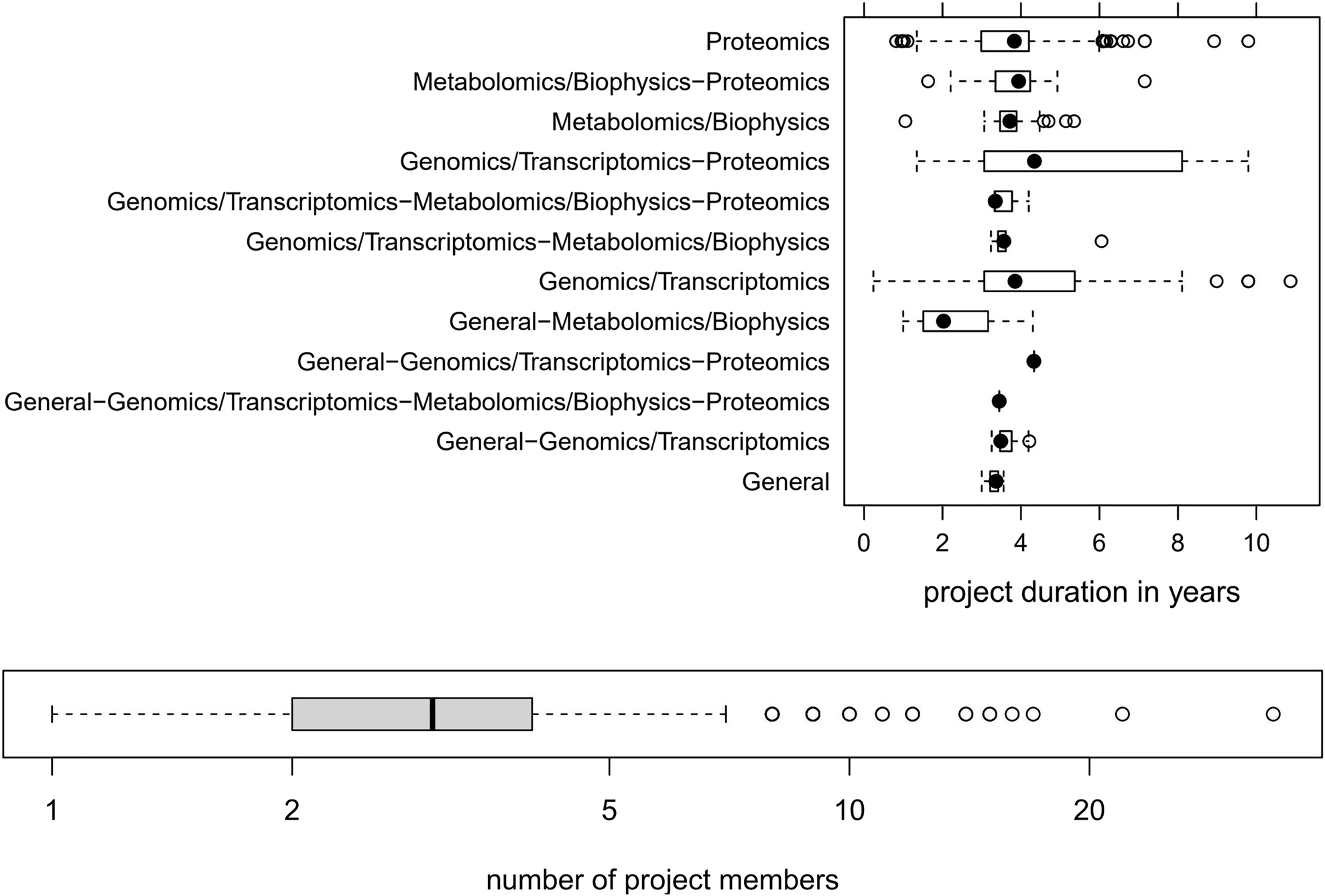
It is important to note that most omics projects run over several years. Figure 2 depicts the typical duration of omics projects and the involved number of project members, as we have witnessed in our core facility over almost two decades.
|
The interdisciplinary nature of such scientific projects requires a powerful and flexible platform for data annotation, communication, and exploration. Information visualization techniques[5][6], especially interactive ones, are well suited to support the exploration phase of the conducted research. However, general support for a wide area of applications is often hampered for different reasons:
- Heterogeneity of structure and missing implementation of standards may disable the easy bundling of existing applications.
- Prototype applications may not be deployable and maintainable in a more extensive system setup, as they may be needed for production use.
- Initial costs per user may be high.[7]
- Scientists usually have limited programming skills.
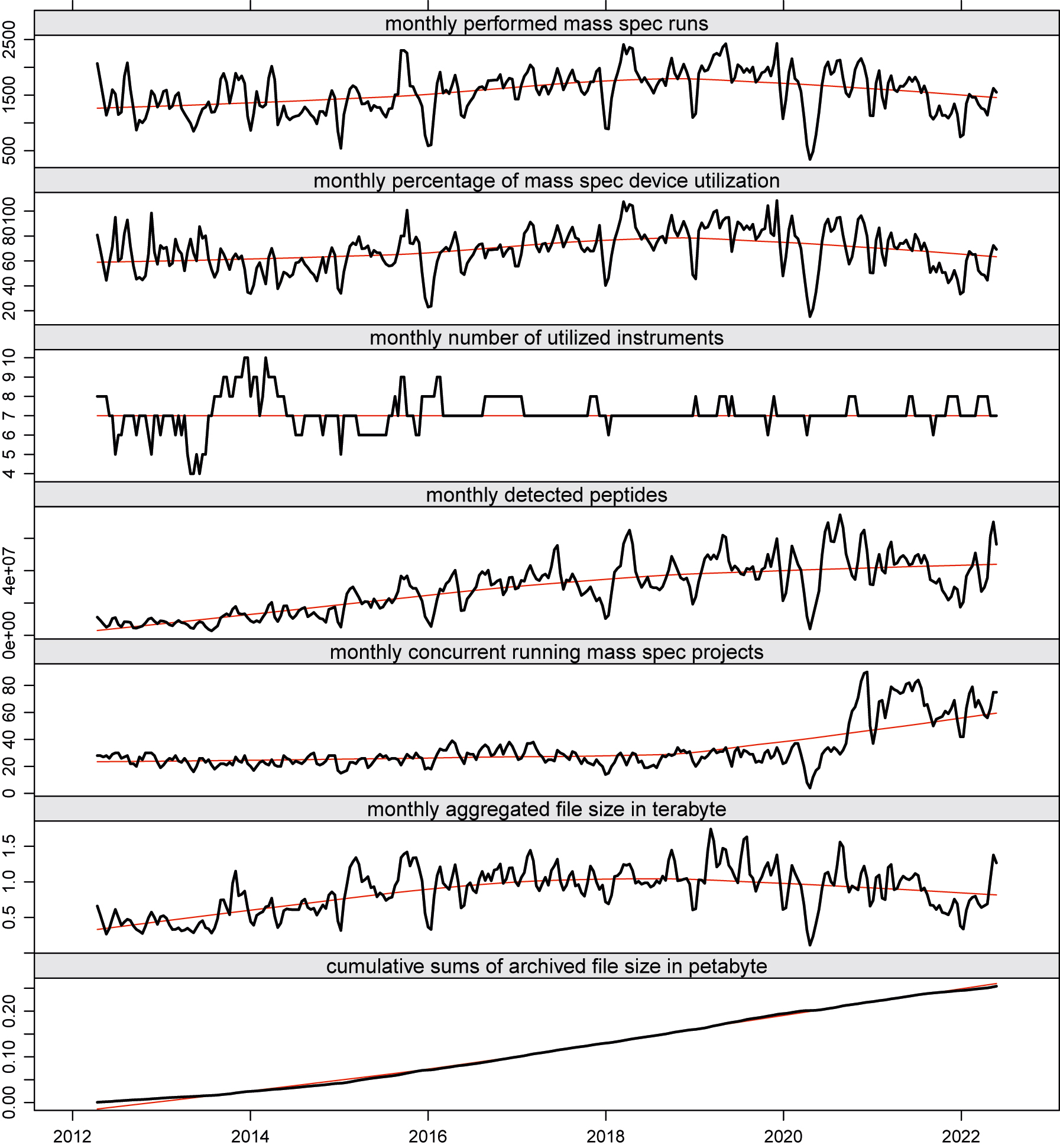
The graphs in Figure 3 depict some key performance indicator (KPI) trends while plotting a monthly sliding window over the aggregated data of conducted runs, the number of concurrent running research projects, and the number of detected peptides (proteins) of our MS unit at the Functional Genomics Center Zurich (FGCZ).
|
Throughout the years, we observed the phenomenon that traditional core IT development cannot cope with the pace of the evolution cycles of data analysis applications.[2][8] Sometimes, new data analysis tools become obsolete before being fully integrated into the core IT system. Consequently, the core IT development team needs to carefully evaluate the latest trends in data analysis and assess which ones should be implemented first, considering not only scientific but also economic factors. At our core facility, we are faced with fundamental data processing and analysis problems due to challenging data formats used in the different scientific areas. In the following, we provide a brief example on a default proteomics workflow, as sketched in Figure 1, but note that the system analogously serves for other technology areas at our facility, such as sequencing, genomics, metabolomics, and single cell analysis.
Our use case
In MS, for example, the mass charge ratio of ions is determined and the data is recorded as a spectrum consisting of a mass–ion charge ratio (m/Z) axis (x coordinates) and an intensity axis (y coordinates). In proteomics— where “everything in life is driven by proteins”[9]—the ions of interest are generated from peptides separated by liquid chromatography (LC) in time (from 30 minutes up to several hours) and recorded with high scan speed (up to 40 Hz). These quickly result in datasets of thousands of scans, each containing several hundreds of ions. Additional complexity is added to the data by the fact that the spectra can be recorded on different MS levels. On MS level 1 (MS1), a simple survey is performed, while on MS level 2 (MS2), a specific ion from an MS1 scan is selected, isolated, fragmented, and the resulting fragment ions are recorded. On MS level 3 (MS3), an ion from an MS2 scan is selected for further fragmentation. To make matters more complicated, on MS2 and higher, different types of activation techniques can be selected for fragmentation. This setup allows for the generation of complex experiments, which in turn results in detailed nested data. To answer a scientific question with such MS data, it is necessary to filter out some selected ions of interest from several million of ions distributed over the data structure. Some typical questions in this area are:
- Which proteins can be identified in an organism under which conditions?
- Can particular post-translational modifications (PTMs), e.g., phosphorylation on serine, threonine, and tyrosine (STY) amino acids, be detected or new ones discovered?
- How do protein abundance levels or PTM patterns change under different conditions?
Ideally, data is measured, automatically transferred to storage, and fed into a meta-database using automated robots in a large-scale high-throughput manner by using proprietary software provided by the vendors, or free software packages. The analysis part is often implemented by using environments such as the Comprehensive R Archive Network (CRAN)[10] and Bioconductor[11][12][13], and packages like shiny[14], allowing the integration of largely known bioinformatics tools[15][16][17][18][19][20] and visualization techniques.[5][6][11][21][22][23] The packages knitr[24] and rmarkdown[25] facilitate automated and reproducible report generation. The art is found in bringing all these tools together to document the research steps such that the data involved can be provided and interpreted correctly. For that, an integrative platform is needed.
Contributions of this work
It is essential to note this paper focuses neither on presenting a new visualization tool nor on introducing a new integrative platform. The goal is to share experiences we made in quickly developing visualization applications in the area of life sciences based on an integrative platform on which we have successfully implemented and run for more than 10 years at our core facility. We will demonstrate how to couple and run real visualization applications on our generic integrative platform, called B-Fabric. As an example, we have chosen the visualization of MS data. Here, we will show how existing software (an R package, e.g., PTM MarkerFinder[26]) can be integrated into a shiny framework. The B-Fabric platform will only be sketched at a level needed to understand this paper’s message. We refer the reader to various works of Türker et al.[8][27][28] for more details on B-Fabric. In the remainder of this paper, we will show how to link applications to B-Fabric to detect a given set of mass patterns by applying the full bandwidth of information visualization techniques developed in the last 30 years and available through environments like R shiny.[14] While this particular application focuses on identifying PTM signatures[26][29], the application itself could be exchanged easily with others. Also, traditional visualization systems, such as Spotfire[30], can be used on the higher layer of our architecture.
The paper organization is as follows. The next section provides a brief overview of the data management system, while two typical application examples—one interactive and one with a graphical user interface—are described after that. The final sections provide some lessons learned and conclusions made with the described solution.
Integrative system platform
Core facilities require a data management and processing platform to cope with the massive data and query sizes of today’s research, as well as with the immense dynamics of evolving analysis methods and tools that can emerge and disappear at a barely perceptible pace. A successful solution has to separate the different concerns levels and avoid bottlenecks caused by a limited number of system programmers and data analysis experts, as well as by restricted storage and computing resources. Concerning data management, it is essential to split the metadata from the actual data content.
Typical system solutions for life sciences research only focus on specific tasks. For instance, analysis tools such as TIBCO Spotfire[30] or SBEAMS[31], myProMS[32], or PeakForest[33] support data preparation and discovery features that help to shape, enrich, and transform life sciences data. Analytics platforms such as KNIME[34], TeraData[35], or GenomeSpace[16] concentrate on enabling users to leverage and pipeline their preferred tools across multiple data types. Tools like the CLC Genomics Workbench[36], PanoramaWeb[37], or Google Genomics mainly provide technologies to analyze, visualize, and share life sciences data. Systems like Seven Bridges[38] go a step further by including basic project and sample management features to organize and share imported and processed data in a more fine-grained, access-controlled fashion. However, to the best of our knowledge, there is no single software that supports the entire needs of a core facility, namely administrative tasks such as managing all required information about the users (researchers), projects, orders, computers, instruments, storage, services, instrument reservations, project reviews, orders, and bookings, as well as analytic tasks such as capturing meta-information about the samples (including the generated raw and post-processed data) and tracking the entire data processing workflow.
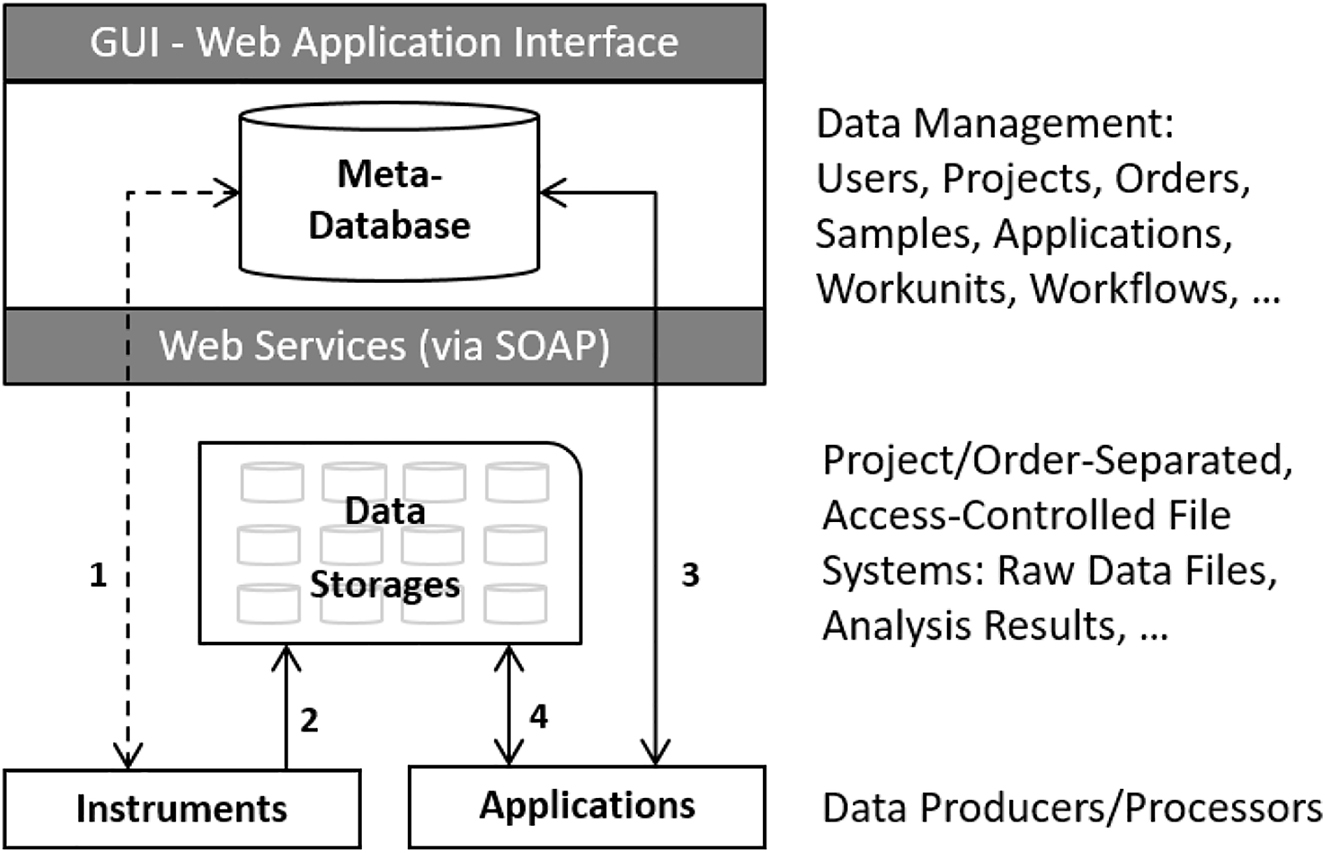
At the FGCZ, we have developed a system called B-Fabric[28], which addresses all these requirements, after having originally started back in 2005 in the same direction as the above-mentioned tools.[27] In the following, we will very roughly sketch the architecture of this system (see Figure 4) without going into further details beyond what is needed to understand the usage of such a system for instant data visualizations and exploration.
|
In the B-Fabric system architecture, experimental data is produced by instruments and applications. This data typically resides on different data storage. The knowledge about this data, in particular about its location, is tracked in the meta-database. Instruments and applications use web services to query and feed the meta-database. As such, all relevant structured data is linked together. As Figure 4 demonstrates, there are four areas of linkage to examine with B-Fabric:
- Data production and generation is completely decoupled from the overall system. Data is produced, for instance, by exchangeable instruments, which usually come with their own storage. Data producers can easily be registered to the overall system. Instruments experts work on this level to install and configure the instruments as needed, as well as to register them in the meta-database to couple them to the overall system.
- Data beamers, which are special applications running on the instrument’s storage, de-multiplex and transfer the produced data into authorized storage. These data storage areas are strictly organized following commonly agreed upon rules. For instance, the data is stored in project-separated file folders, so-called containers, representing the access units for all data produced in a project. At this level, bioinformaticians, who are responsible for the maintenance of the different storage areas, are also responsible for implementing the data beamers based on their knowledge of the corresponding storage areas.
- The meta-database is the brain of the entire system[8] which holds all the knowledge relevant to the core facility. It documents the projects and orders of all users. It stores the samples processed in these projects and orders, as well as the relevant information about the applied procedures and workflows. Thanks to the applications, the meta-database also holds all necessary information about the produced data files and thus provide a homogenous overall picture over highly heterogeneous and autonomous data storage areas. Among other things, it allows for the questions of “Who created which data in which project, and with which samples and with which protocols?” An advanced role model implements a fine-grained access control such that the users can only see the piece of the data world that they are allowed to. The data of the meta-database is accessible via graphical user interfaces (GUI) as well as web services. The entity concepts are realized in such a general manner that the meta-database is prepared to store any type of future research data.
- External applications exploit the metadata to access, process, and visualize the produced research data. The required data preprocessing, including the data crunching, is usually performed on external infrastructures (clusters/grids/clouds[39]). Visualization software, such as the shiny server[14], is an example of the implementation of an external application. The required data is fetched via the web services of the underlying infrastructure. Web services can register the analysis result in the meta-database if needed.
Essential for this evolutionary architecture is implementing a highly generic application concept, which allows for implementing dynamic, ad-hoc workflows of any type to keep track of the entirety of data processing. Using preceding and succeeding relationships, all applications know which data they can work on.[8]
The current B-Fabric 11 release is built upon Jakarta EE, JSF, PrimeFaces, OmniFaces (web application development), PostgreSQL (database management), Apache Lucene (full text search), and SOAP (web service). B-Fabric has run in daily business at the FGCZ since February 2007. Table 1 lists some numbers showing its usage in our three technology platforms: Genomics, Metabolomics, and Proteomics.
| ||||||||||||||||||||||||||||
Application
In this section, we exemplarily demonstrate how our platform is used for ad-hoc linking a visualization software solution to our system in a way that the entire user community can benefit from it. We use the R environment in the example below, but the interface described in this manuscript can be used by any other programming environment supporting Web Services Description Language (WSDL). While the first application example attaches a higher-level interactive user interface to the data management system, the second example is more of a technical nature to demonstrate how the application programmer can glue together data, metadata, and visualization libraries on the R command line. Since we focus more on the technical description in the text, the caption will give more details on what is displayed on the visualization in Figures 6 and 7.
Setup and configuration
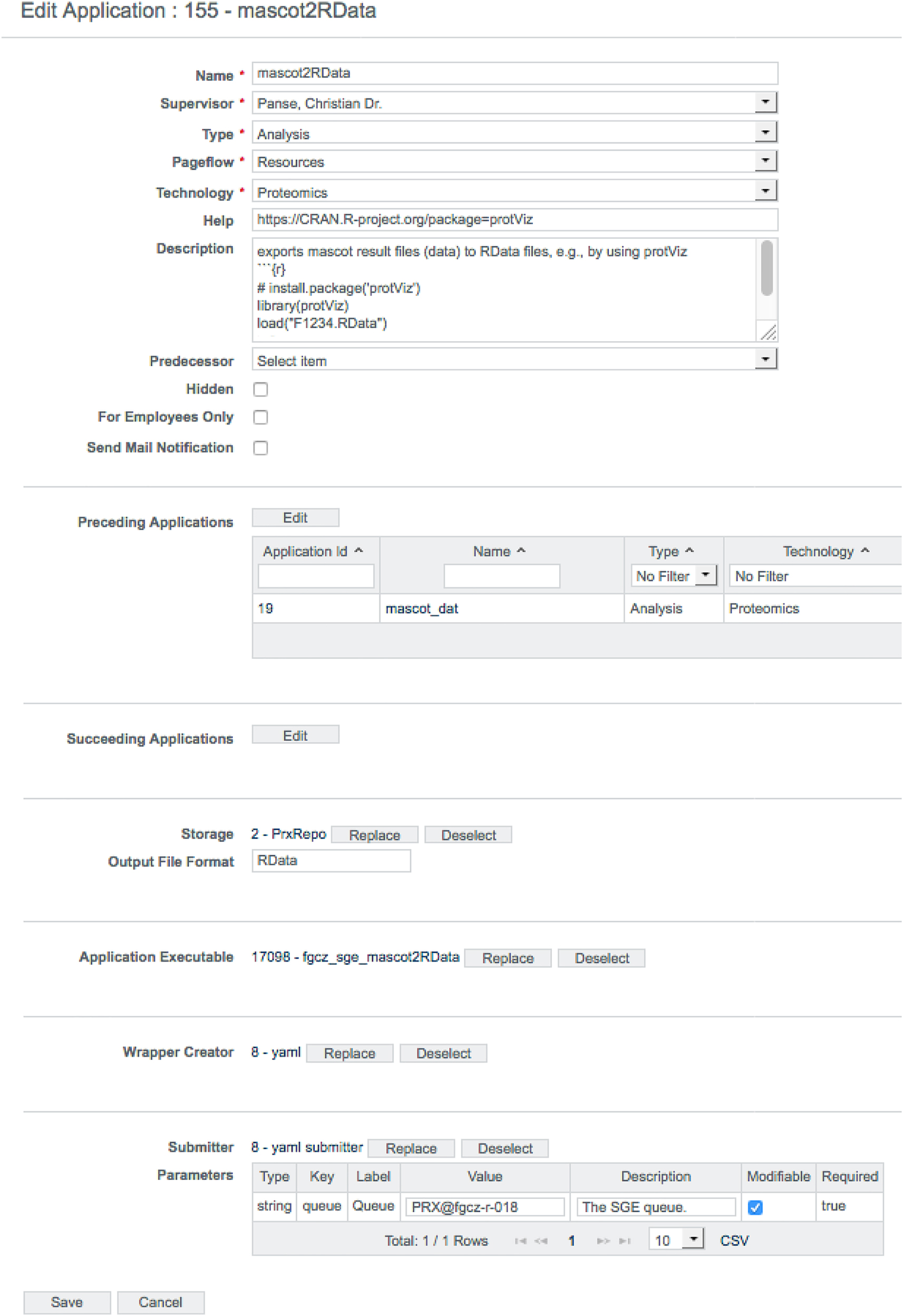
Before an application can be applied to the data, it has to be configured properly. The B-Fabric platform not only allows the annotation of metadata but also provides an annotation mechanism for maintaining the application configuration. As shown in Figure 5, B-Fabric allows registering applications with their specific properties, e.g., type or technology. The tremendous amount of possible flexibility is achieved by attaching executables written externally in any language and allowing for the wrapping of this code by a previously registered wrapper creator, which provides the abstraction for feeding the application executable with the needed metadata. In addition, the submitter abstraction allows running the application executable on a configured computation environment of choice. Every application takes an input (in that context, it is also an application) and provides an output. The field "Preceding Applications" defines the application’s input, and ad-hoc workflows become a reality. Application ID 155, shown in Figure 5, requires as input a peptide identification search[15] application having an application ID of 19. Depending on the computational need, the jobs can be performed on a local compute-server or an attached cloud infrastructure (see section four of Aleksiev et al.[39]). The wrapper creator mechanism implements the job staging. The XML below (Listing 1 and 2) defines the environment where the application is executed and what script or binary file is used. It is also possible to define a list of arguments and pre-defined argument values to be passed to the application executable. The XML form for the executable configuration turned out to be easier to maintain and deploy, especially if the application requires a long list of user parameters.
|
|
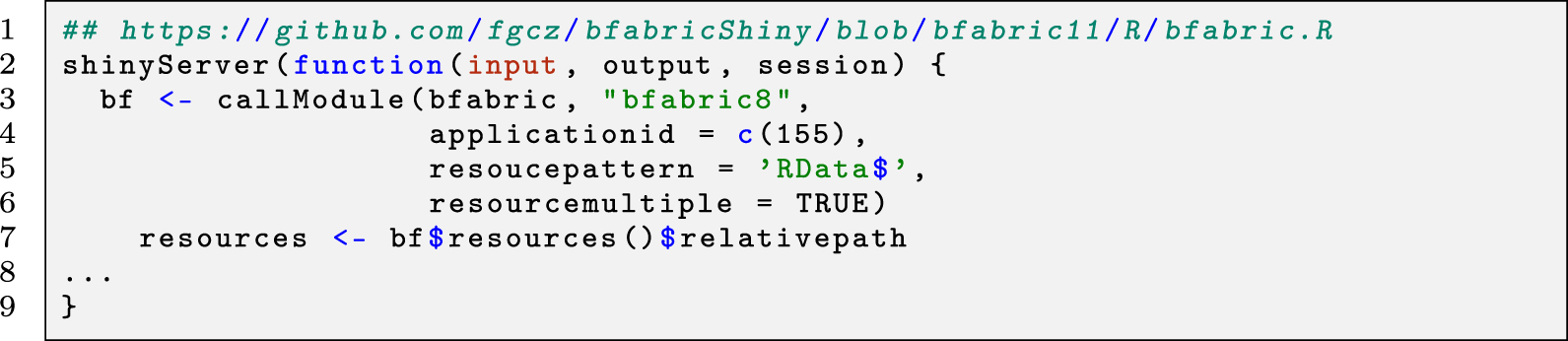
In the example using R shiny, all jobs created by that application (ID = 155) are available (given the user is authorized) in the platform’s user interface, e.g., by calling the module as shown in the following tiny code snippet (Listing 2).
|
The retrieved metadata (e.g., sample information, storage, and the file path of each selected resource) are kept in the bf object (line 6). This object can later be used as a container for any programming purpose.
Interactive visualization
To provide a one-to-one example for the generic description of the platform, sketched in Figure 4, we refer to each item number of that figure.
- The recording of the raw data on today’s MS device can easily take up several days depending on the length of the liquid chromatography–tandem mass spectrometry (LC-MS/MS) runs, the number of samples and the number of quality control (QC) runs scheduled by the systems expert. During data recording, the data is saved onto an instrument control computer. After completion, the data is automatically copied to data storage by robots, which are responsible for the data transfer between instruments and data storage.
- A local copy of the data will remain as a backup for two weeks on the instrument computer before deletion. A data storage is typically a storage area network (SAN) and is attached to the data producing machines and the computing area through an optical fiber network. XFS turned out to be the file system of choice. Directly after synchronization of the data to the SAN (see arrow 2 in Figure 4), a first preprocessing will be triggered, converting the proprietary raw file format into a generic machine-readable format, e.g., Mascot generic file (mgf) or mzXML. Ideally, the annotation (see arrow 1 in Figure 4) in the platform has already taken place prior to the data analysis, but it needs to be completed at last when the raw data is “imported” into the meta-database where the corresponding raw data will be linked.
- At this next step, the systems experts or scientists trigger a so-called “MS/MS database search,” e.g., by using Matrix Science Mascot Server[15] or comet.[40] Those search algorithms assign the MS data (spectra) to protein sequences. The amino acid sequences were predicted by using DNA/RNA sequences. A quantitative measure is derived from the area under the curve (AUC) of the ion counts signals over the retention time. For further processing, this data is usually converted into an exchange format, usually XML (see also HUPO Proteomics Standards Initiative[41]). An XML conversion of such data has the drawback that the resulting files are very large, and the parsing is very time-consuming. As a practical step, the XML is further processed into a more typical R list, where, if necessary, some filtering can already be applied and saved as a compressed RData “container.”[42] Similar libraries and procedures are also available for the programming languages Python, Ruby, and Perl. After this step, the data can be read directly into the application.
- A system-specific R shiny module[43] is handling all the authentication and data staging tasks (see arrows 3 and 4 in Figure 4) using the web services interface of the B-Fabric platform. The data staging is performed by using the network file system (NFS, RFC7530) or the secure shell (SSH, RFC4254) protocols. Also, for mobile computers, a proxy solution is possible.
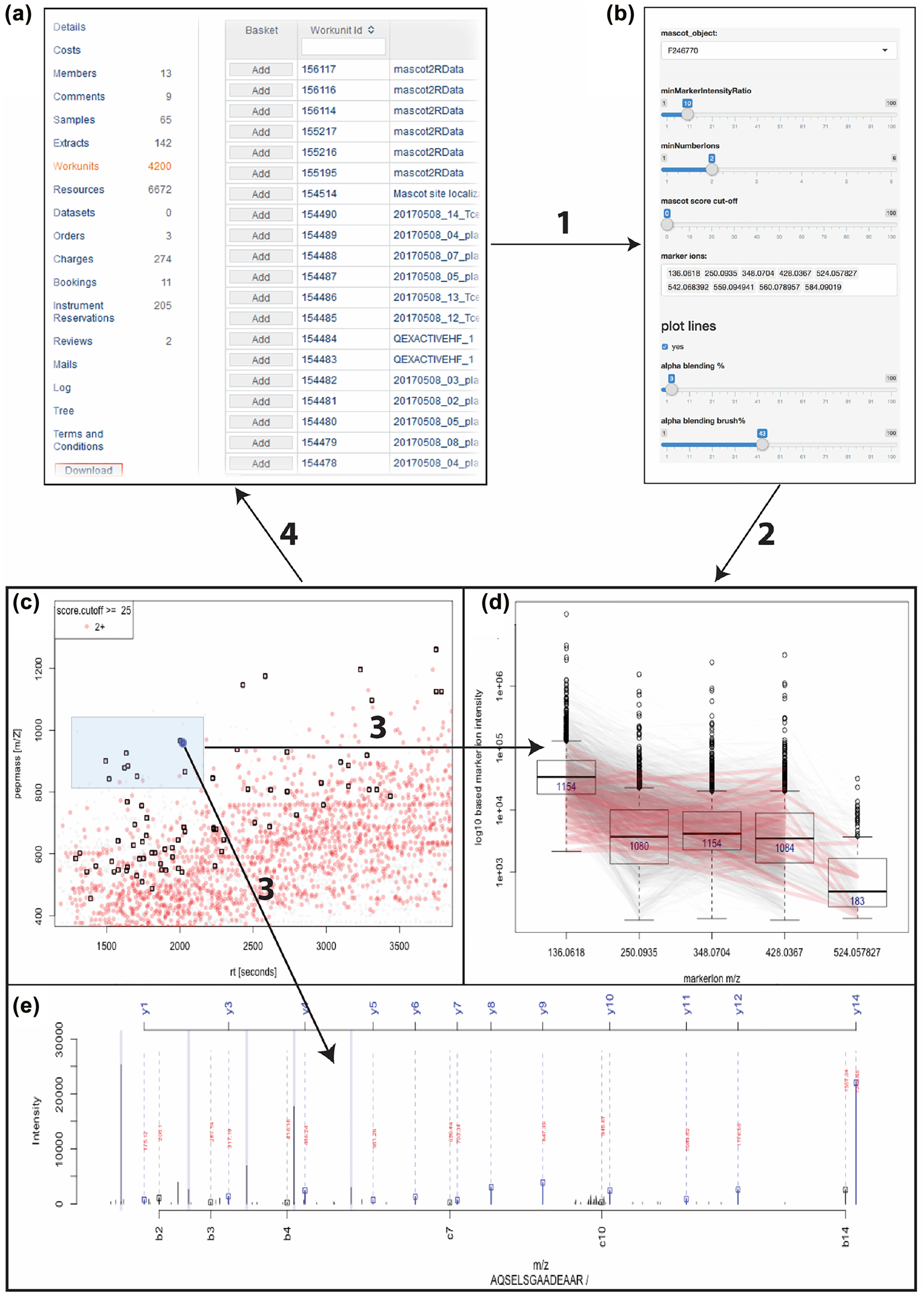
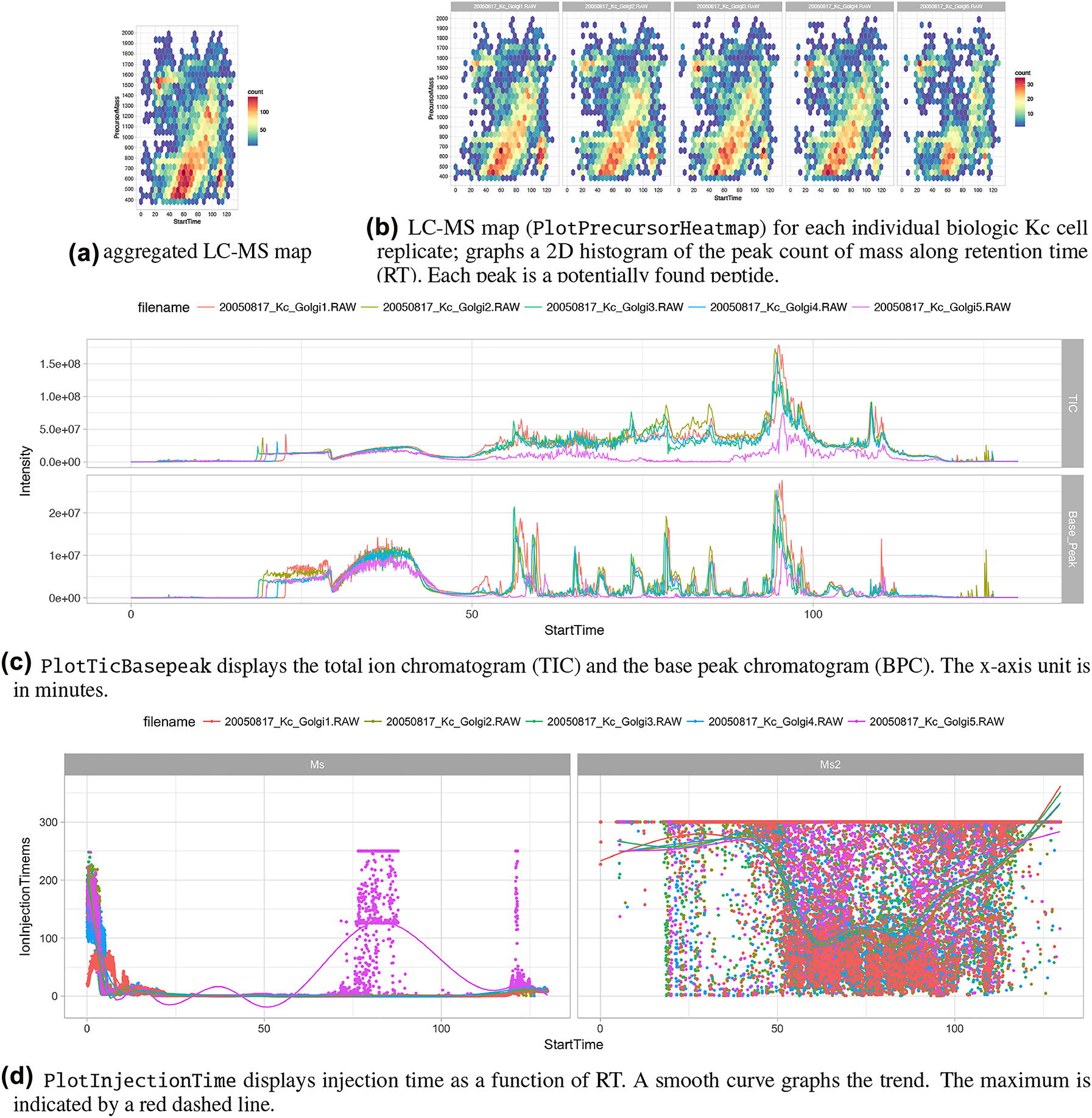
The screenshots in Figure 6 display the first application example with the B-Fabric platform (see also arrow 3 in Figure 4). The application software[15][26][29][44] itself detects and quantifies known marker ions released during the different types of ion fragmentation of the peptide during the data acquisition. Together with the findings of the previously mentioned “MS/MS database search” and the dynamic graphical analysis, it is a powerful tool for the decision-making exploration process of the scientists.
|
Figure 6 is outlined a such. (a) The screenshot gives an overview of the research project statistics, e.g., the number of samples and measurements, by querying the meta-database. Particular resources can be collected into an “Amazon” basket and transferred as input to related applications. (b) The panel displays some relevant parameter settings for the selected application. Most important are the input containing the list of the mass pattern. (c) Each data item represents a picked peptide candidate signal on the m/Z versus retention time zoomed-in scatterplot of one LC-MS instrument run. Color represents the charge state (filtered; only 2+). The black boxes identify signals where the expected ADPr fragment ion mass pattern was detected using the PTM MarkerFinder package.[26] (d) The boxplots display the distribution of the ion count of each tandem mass fragmentation ion pattern . The lines connecting each data item on the boxplot depict the trend. Brushing, the red colored lines selected by the blue box in (c), enable the “discovery mode” of the application. (e) Using linking, the peptide-spectrum match is shown. The plot shows that the highest peaks are assigned to the b and y ions. Additionally, the marker ions are highlighted (as QC). The result of the exploration process and the used parameters can be made persistent by feeding it back to the meta-database and the data storage.
Command-line-triggered visual exploration
The second application example illustrates the interaction between an application (performing QC) and the B-Fabric platform on the command line level using the R environment. On the one hand, it shows how a more skilled user of programming can use the platform interface. On the other hand, it also demonstrates how a more complex visualization application can be implemented. Of note, the workflow consists of steps (3) and (4) only (see Figure 4). The depicted code snippet in Listing 3 requires an authentication configuration, saved in login and webservicepassword in line 11, which is used for accessing the B-Fabric platform.
|
The example screens a smaller dataset generated as part of an experiment to identify the entire fruit fly model organism.[4] Lines 11–13 perform a database query via WSDL to the B-Fabric platform. This particular query returns all resources belonging to a given workunitid=165473. A workunit is a collection of files. For a more complex example like an expression analysis, we could also retrieve additional metadata, e.g., the sample annotation and treatment for each measurement. Besides the read mechanism, the webservice methods save, and delete can be used to save or delete the result of an analysis. By using the save method also, intermediate result files can be uploaded to B-Fabric. Lines 15–16 concatenate the root directory with each resource’s extracted relative file path. For our example application, to read the content of the proprietary instrument files and visualize it, we use an R package rawDiag[45], a software tool supporting rational method optimization by providing MS operator-tailored diagnostic plots of scan level metadata. Lines 19–21 read the proprietary MS-formatted data in parallel. This method can provide the mechanism of retrieving the data via remote method invocation (RMI) or using the Secure Shell Protocol (SSH). The idea is to avoid copying the entire file.
From line 24 on, we generate the graphics displayed in Figure 7. The graphics on Figure 7 (b) can also be used to perform visual representation as a set of thumbnails of a bigger dataset.[46] The design of generating the visualizations follows the concept described by Wilkinson [22] using ggplot2.[47] The rawDiag package[45] can be used stand-alone or plugged into the B-Fabric system (see http://fgcz-ms-shiny.uzh.ch:8080/bfabric_rawDiag/ by using the bfabricShiny module.[43] It is used as an application running in a web browser and from the R[10] command line.
|
Furthermore, as input for Figure 7, we use data measured on a Thermo Finnigan LTQ device (2005) using a 96-minute LC gradient. Injected were typically digested Golgi Kc cell line replicates derived from a fruit fly (D. melanogaster)[4] (see also Figure 1(a)). Figure 7(a) graphs an aggregated “overview map” of the LC-MS runs of the five replicates. The 2D map shows a typical pattern, which means after a loading phase (15 minutes), lighter peptides start to elute, followed by heavier ones. Figure 7(b) graphs individual LC-MS runs. Replicate five, 20050817_Kc_Golgi5, shows a different pattern after 50 minutes. Figure 7(c) shows that total ion count (TIC) drops after 50 minutes (pink curve). In Figure 7(d), the injection time plot graphs the time used for filling the ion trap of the MS device for the scans of MS level 1 (left) and level (2) right. At the latest, seeing the point cloud after 60 minutes on the graph (d) indicates a malfunction often caused by the spray attached to the ion source.
Lessons learned
Deploying the newest visualization and exploration solutions to a productive environment is challenging. The described solution based on using the B-Fabric platform enables the linking of various visualization, data processing, and data analysis software tools to a data management system containing massive repositories of life sciences data and metadata, such as definitions of applied experimental designs. The platform analyzes massive amounts of genomics, sequencing, proteomics, and metabolomics data generated at our core facility for over a decade. Over the years, we witnessed the rapid evolution of technologies in the various life sciences areas, which consequently demands an integrative platform ready to couple with any potential future application with little effort.
Concretely, we learned the following lessons:
1. Every institution has its superior solution. Concerning core facilities, the systems are often adapted to the internal business needs, for instance, reviewing, accounting, ordering, and all other required administrative processes. Other facilities usually rely on commercial laboratory information management systems (LIMS). Once a LIMS is in operation, it is almost impossible to exchange such a system due to the enormous financial and human capital investment. Usually, LIMS are implemented for a specific domain and are not generic enough. From the software engineering point of view, it is essential to have a generic platform providing an interface to read and save any type of data.
2. Follow commonly defined standards.' Sticking to standards in packaging analysis software eases the deployment and teaching of the software packages. Applications for statistical modeling, differential expression analysis, p-value adaption, data mining, and machine learning, e.g., data imputation missing values and information visualization techniques. Platforms such as Bioconductor[48][49] have already provided solutions for more than a decade, but the deployment and the provisioning of data and metadata (containing the experimental design) to the scientist having the domain knowledge remain unsolved.
3. Divide and conquer through decoupling. In an early phase, more than 10 years ago, our system was designed as one monolithic stand-alone system where all applications were integrated into a platform with one programming language. As a result, the software engineering tasks were distributed over a few shoulders (software engineers). Consequently, releasing small changes in an analysis module took too long. The decoupling of the data management and the application part led to the decoupling and full parallelization of the development cycles. Software engineering groups and statisticians/bioinformatics groups develop with different release times, using different development tools and programs in different languages. As an additional benefit of the separation, the analysis tools can also be deployed on a different site running on different systems, which leads to more robustness of the code, allows better reproducibility, and finally enables more transparency to the R research. Good examples of pluggable visualization are found in PTM MarkerFinder[26] and rawDiag[45]; others examples are found in specL[12], NestLink[50], and prolfqua.[51]
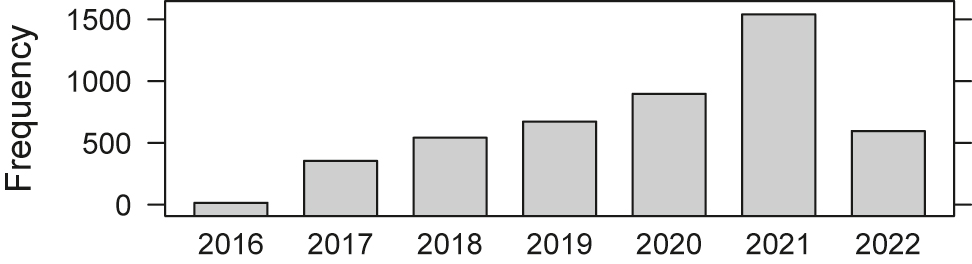
Since we introduced the described concept in 2016, we have seen a significant increment in the number of executed applications. Note, however, that we only counted if the result of an application is stored (see Figure 8).
|
4. Eliminate time-consuming user interaction by increasing user satisfaction. Detect and fix issues before a large group of users recognizes them and creates several tickets for the same issue. As such, you increase user satisfaction and avoid time-consuming communication. Also, a common, intuitive interface saves time for training and code adaption.
5. Practice sustainability through consistent metadata annotation. An essential requirement for effective data analysis is a high-quality metadata annotation encoding the experimental design. This is true even when, as in our case, project duration typically last two to five years (as shown in Figure 2). For instance, the best visualization tool/technique is worthless if the visualized data cannot be linked to the corresponding metadata, which builds the fundament for the correct interpretation of the visualized data.
Conclusions
In this paper, we have sketched how ad-hoc visualizations and analytics can be realized on top of an integrative platform that captures and processes any kind of data and scientific processes. For that, we were using our integrative B-Fabric platform. The key idea behind the platform is to decouple metadata management from data processing and visualization components. The advantage is that development in the different areas can take place at different speeds, and each area can benefit from using specific software environments, which provide the basis for realizing optimal solutions in the corresponding areas. The two chosen implemented application examples illustrated how the visualization tools could interface in such a platform. The evolutionary B-Fabric platform has already proven its capability in daily business for more than a decade, especially by providing interactive visual data exploration solutions in the omics areas to a broader class of users. The platform enables ad-hoc attaching visualization tools and makes them available to a vast set of annotated data and an entire user community. Since parameter and intermediate results can be instantly made persistent, the platform is also a milestone towards the trend of reproducible research. It provides the fundament of a maintainable and robust software stack that is not limited only to the area of life sciences. Furthermore, in this way, B-Fabric helps scientists to meet requirements from funding agencies, journals, and academic institutions to publish data in a findable, accessible, interoperable, and reusable (FAIR) way.[52]
Abbreviations, acronyms, and initialisms
- ADP: adenosine diphosphate
- CRAN: Comprehensive R Archive Network
- FAIR: findable, accessible, interoperable, and reusable
- FGCZ: Functional Genomics Center Zurich
- GUI: graphical user interface
- KPI: key performance indicator
- LC: liquid chromatography
- LT: linear ion trap MS
- m/Z: mass–ion charge ratio
- MS: mass spectrometry
- MS/MS: tandem mass spectrometry
- PTM: post-translational modification
- QC: quality control
- RMI: remote method invocation
- SSH: Secure Shell Protocol
- STY: serine, threonine, and tyrosine
- TIC: total ion count
- WSDL: Web Services Description Language
Acknowledgements
B-Fabric would not exist in its current state without the support and the continuous commitment of the FGCZ management, the patience of all the users of the FGCZ UZH|ETHZ community, the desire for new features of our colleagues, and the never-ending enthusiasm and creativity of all past and current B-Fabric developers.
Author contributions
All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
Research funding
None declared.
System and data availability
All R packages[45][50][53][54], including documentation, unit tests and examples, are available under the GNU General Public License v3.0 via the following URLs:
- https://github.com/fgcz/rawDiag
- https://bioconductor.org/packages/rawrr/
- https://bioconductor.org/packages/NestLink/
- https://github.com/fgcz/bfabricShiny
- https://CRAN.R-project.org/package=protViz
The B-Fabric data management platform is available for all registered users of the FGCZ through https://fgcz-bfabric.uzh.ch. The code of B-Fabric is closed since it is commercially licensed. Therefore, we restricted the presentation of the platform to a rough level that is needed to understand how omics workflows can be realized with an integrative platform such as B-Fabric. Interested parties are welcome to approach Can Türker.
Conflict of interest
The authors declare no conflicts of interest regarding this article.
References
- ↑ "Definitions". Research Core Facilities at Drexel University. Drexel University. 2023. https://drexel.edu/core-facilities/resources/definitions/.
- ↑ 2.0 2.1 Barkow-Oesterreicher, Simon; Türker, Can; Panse, Christian (1 December 2013). "FCC – An automated rule-based processing tool for life science data" (in en). Source Code for Biology and Medicine 8 (1): 3. doi:10.1186/1751-0473-8-3. ISSN 1751-0473. PMC PMC3614436. PMID 23311610. https://scfbm.biomedcentral.com/articles/10.1186/1751-0473-8-3.
- ↑ Chiva, Cristina; Mendes Maia, Teresa; Panse, Christian; Stejskal, Karel; Douché, Thibaut; Matondo, Mariette; Loew, Damarys; Helm, Dominic et al. (4 June 2021). "Quality standards in proteomics research facilities: Common standards and quality procedures are essential for proteomics facilities and their users" (in en). EMBO reports 22 (6). doi:10.15252/embr.202152626. ISSN 1469-221X. PMC PMC8183401. PMID 34009726. https://onlinelibrary.wiley.com/doi/10.15252/embr.202152626.
- ↑ 4.0 4.1 4.2 Brunner, Erich; Ahrens, Christian H; Mohanty, Sonali; Baetschmann, Hansruedi; Loevenich, Sandra; Potthast, Frank; Deutsch, Eric W; Panse, Christian et al. (1 May 2007). "A high-quality catalog of the Drosophila melanogaster proteome" (in en). Nature Biotechnology 25 (5): 576–583. doi:10.1038/nbt1300. ISSN 1087-0156. http://www.nature.com/articles/nbt1300.
- ↑ 5.0 5.1 Becker, Richard A.; Cleveland, William S.; Wilks, Allan R. (1 November 1987). "Dynamic Graphics for Data Analysis". Statistical Science 2 (4). doi:10.1214/ss/1177013104. ISSN 0883-4237. https://projecteuclid.org/journals/statistical-science/volume-2/issue-4/Dynamic-Graphics-for-Data-Analysis/10.1214/ss/1177013104.full.
- ↑ 6.0 6.1 Keim, D.A. (Jan.-March/2002). "Information visualization and visual data mining". IEEE Transactions on Visualization and Computer Graphics 8 (1): 1–8. doi:10.1109/2945.981847. http://ieeexplore.ieee.org/document/981847/.
- ↑ van Wijk, J.J. (2005). "The Value of Visualization". VIS 05. IEEE Visualization, 2005. (Minneapolis, MN, USA: IEEE): 79–86. doi:10.1109/VISUAL.2005.1532781. ISBN 978-0-7803-9462-9. http://ieeexplore.ieee.org/document/1532781/.
- ↑ 8.0 8.1 8.2 8.3 Türker, Can; Akal, Fuat; Joho, Dieter; Panse, Christian; Barkow-Oesterreicher, Simon; Rehrauer, Hubert; Schlapbach, Ralph (22 March 2010). "B-Fabric: the Swiss Army Knife for life sciences" (in en). Proceedings of the 13th International Conference on Extending Database Technology (Lausanne Switzerland: ACM): 717–720. doi:10.1145/1739041.1739135. ISBN 978-1-60558-945-9. https://dl.acm.org/doi/10.1145/1739041.1739135.
- ↑ Gonick, Larry; Wheelis, Mark (2007). The cartoon guide to genetics (Updated ed., 40. [print,], 1st Collins ed ed.). New York, NY: Collins Reference. ISBN 978-0-06-273099-2.
- ↑ 10.0 10.1 "The Comprehensive R Archive Network". Institute for Statistics and Mathematics of WU. 2022. https://cran.r-project.org/.
- ↑ 11.0 11.1 Gatto, Laurent; Breckels, Lisa M.; Naake, Thomas; Gibb, Sebastian (1 April 2015). "Visualization of proteomics data using R and Bioconductor" (in en). PROTEOMICS 15 (8): 1375–1389. doi:10.1002/pmic.201400392. ISSN 1615-9853. PMC PMC4510819. PMID 25690415. https://onlinelibrary.wiley.com/doi/10.1002/pmic.201400392.
- ↑ 12.0 12.1 Panse, Christian; Trachsel, Christian; Grossmann, Jonas; Schlapbach, Ralph (1 July 2015). "specL—an R/Bioconductor package to prepare peptide spectrum matches for use in targeted proteomics" (in en). Bioinformatics 31 (13): 2228–2231. doi:10.1093/bioinformatics/btv105. ISSN 1367-4811. https://academic.oup.com/bioinformatics/article/31/13/2228/195976.
- ↑ Kockmann, Tobias; Trachsel, Christian; Panse, Christian; Wahlander, Asa; Selevsek, Nathalie; Grossmann, Jonas; Wolski, Witold E.; Schlapbach, Ralph (1 August 2016). "Targeted proteomics coming of age - SRM, PRM and DIA performance evaluated from a core facility perspective" (in en). PROTEOMICS 16 (15-16): 2183–2192. doi:10.1002/pmic.201500502. https://onlinelibrary.wiley.com/doi/10.1002/pmic.201500502.
- ↑ 14.0 14.1 14.2 Chang, W.; Cheng, J.; Allaire, J.J. et al. (28 March 2016). "shiny: Web Application Framework for R (0.13.2)". CRAN. Institute for Statistics and Mathematics of WU. https://cran.r-project.org/package=shiny.
- ↑ 15.0 15.1 15.2 15.3 Perkins, D. N.; Pappin, D. J.; Creasy, D. M.; Cottrell, J. S. (1 December 1999). "Probability-based protein identification by searching sequence databases using mass spectrometry data". Electrophoresis 20 (18): 3551–3567. doi:10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. ISSN 0173-0835. PMID 10612281. https://pubmed.ncbi.nlm.nih.gov/10612281.
- ↑ 16.0 16.1 Qu, Kun; Garamszegi, Sara; Wu, Felix; Thorvaldsdottir, Helga; Liefeld, Ted; Ocana, Marco; Borges-Rivera, Diego; Pochet, Nathalie et al. (1 March 2016). "Integrative genomic analysis by interoperation of bioinformatics tools in GenomeSpace" (in en). Nature Methods 13 (3): 245–247. doi:10.1038/nmeth.3732. ISSN 1548-7091. PMC PMC4767623. PMID 26780094. http://www.nature.com/articles/nmeth.3732.
- ↑ Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V. et al. (1 July 2012). "ExPASy: SIB bioinformatics resource portal" (in en). Nucleic Acids Research 40 (W1): W597–W603. doi:10.1093/nar/gks400. ISSN 0305-1048. PMC PMC3394269. PMID 22661580. https://academic.oup.com/nar/article-lookup/doi/10.1093/nar/gks400.
- ↑ Deutsch, Eric W.; Mendoza, Luis; Shteynberg, David; Farrah, Terry; Lam, Henry; Tasman, Natalie; Sun, Zhi; Nilsson, Erik et al. (1 March 2010). "A guided tour of the Trans-Proteomic Pipeline" (in en). PROTEOMICS 10 (6): 1150–1159. doi:10.1002/pmic.200900375. PMC PMC3017125. PMID 20101611. https://onlinelibrary.wiley.com/doi/10.1002/pmic.200900375.
- ↑ Chambers, Matthew C; Maclean, Brendan; Burke, Robert; Amodei, Dario; Ruderman, Daniel L; Neumann, Steffen; Gatto, Laurent; Fischer, Bernd et al. (1 October 2012). "A cross-platform toolkit for mass spectrometry and proteomics" (in en). Nature Biotechnology 30 (10): 918–920. doi:10.1038/nbt.2377. ISSN 1087-0156. PMC PMC3471674. PMID 23051804. http://www.nature.com/articles/nbt.2377.
- ↑ Breckels, Lisa M.; Gibb, Sebastian; Petyuk, Vladislav; Gatto, Laurent (23 November 2016), Bessant, Conrad, ed., "R for Proteomics" (in en), Proteome Informatics (The Royal Society of Chemistry): 321–364, doi:10.1039/9781782626732-00321, ISBN 978-1-78262-428-8, https://books.rsc.org/books/book/629/chapter/311811/R-for-Proteomics. Retrieved 2023-03-13
- ↑ Cleveland, William S. (1993). Visualizing data. Murray Hill, N.J. : [Summit, N.J: At&T Bell Laboratories ; Published by Hobart Press. ISBN 978-0-9634884-0-4.
- ↑ 22.0 22.1 Wilkinson, Leland; Wills, Graham (2005). The grammar of graphics. Statistics and computing (2nd ed ed.). New York: Springer. ISBN 978-0-387-24544-7.
- ↑ Broeksema, Bertjan; McGee, Fintan; Calusinska, Magdalena; Ghoniem, Mohammad (1 October 2014). "Interactive visual support for metagenomic contig binning". 2014 IEEE Conference on Visual Analytics Science and Technology (VAST) (Paris, France: IEEE): 233–234. doi:10.1109/VAST.2014.7042506. ISBN 978-1-4799-6227-3. http://ieeexplore.ieee.org/document/7042506/.
- ↑ Xie, Yihui (19 April 2016) (in en). Dynamic Documents with R and knitr (0 ed.). Chapman and Hall/CRC. doi:10.1201/b15166. ISBN 978-0-429-17103-1. https://www.taylorfrancis.com/books/9781482203547.
- ↑ Allaire, J.J.; Xie, Y.; McPherson, J. et al. (15 June 2017). "rmarkdown: Dynamic Documents for R (1.6)". CRAN. Institute for Statistics and Mathematics of WU. https://cran.r-project.org/package=rmarkdown.
- ↑ 26.0 26.1 26.2 26.3 26.4 Nanni, Paolo; Panse, Christian; Gehrig, Peter; Mueller, Susanne; Grossmann, Jonas; Schlapbach, Ralph (1 August 2013). "PTM MarkerFinder, a software tool to detect and validate spectra from peptides carrying post-translational modifications" (in en). PROTEOMICS 13 (15): 2251–2255. doi:10.1002/pmic.201300036. https://onlinelibrary.wiley.com/doi/10.1002/pmic.201300036.
- ↑ 27.0 27.1 Türker, Can; Stolte, Etzard; Joho, Dieter; Schlapbach, Ralph (2007), Cohen-Boulakia, Sarah; Tannen, Val, eds., "B-Fabric: A Data and Application Integration Framework for Life Sciences Research" (in en), Data Integration in the Life Sciences (Berlin, Heidelberg: Springer Berlin Heidelberg) 4544: 37–47, doi:10.1007/978-3-540-73255-6_6, ISBN 978-3-540-73254-9, http://link.springer.com/10.1007/978-3-540-73255-6_6. Retrieved 2023-03-13
- ↑ 28.0 28.1 Türker, C.; Schmid, M.; Joho, D. et al. (2018). "B-Fabric User Manual". https://fgcz-bfabric.uzh.ch/wiki/B-Fabric+Manual.
- ↑ 29.0 29.1 Gehrig, Peter M.; Nowak, Kathrin; Panse, Christian; Leutert, Mario; Grossmann, Jonas; Schlapbach, Ralph; Hottiger, Michael O. (6 January 2021). "Gas-Phase Fragmentation of ADP-Ribosylated Peptides: Arginine-Specific Side-Chain Losses and Their Implication in Database Searches" (in en). Journal of the American Society for Mass Spectrometry 32 (1): 157–168. doi:10.1021/jasms.0c00040. ISSN 1044-0305. https://pubs.acs.org/doi/10.1021/jasms.0c00040.
- ↑ 30.0 30.1 Ahlberg, Christopher (1 December 1996). "Spotfire: an information exploration environment" (in en). ACM SIGMOD Record 25 (4): 25–29. doi:10.1145/245882.245893. ISSN 0163-5808. https://dl.acm.org/doi/10.1145/245882.245893.
- ↑ Marzolf, Bruz; Deutsch, Eric W.; Moss, Patrick; Campbell, David; Johnson, Michael H.; Galitski, Timothy (6 June 2006). "SBEAMS-Microarray: database software supporting genomic expression analyses for systems biology". BMC bioinformatics 7: 286. doi:10.1186/1471-2105-7-286. ISSN 1471-2105. PMC 1524999. PMID 16756676. https://pubmed.ncbi.nlm.nih.gov/16756676.
- ↑ Poullet, Patrick; Carpentier, Sabrina; Barillot, Emmanuel (1 August 2007). "myProMS, a web server for management and validation of mass spectrometry-based proteomic data" (in en). PROTEOMICS 7 (15): 2553–2556. doi:10.1002/pmic.200600784. https://onlinelibrary.wiley.com/doi/10.1002/pmic.200600784.
- ↑ Paulhe, Nils; Canlet, Cécile; Damont, Annelaure; Peyriga, Lindsay; Durand, Stéphanie; Deborde, Catherine; Alves, Sandra; Bernillon, Stephane et al. (1 June 2022). "PeakForest: a multi-platform digital infrastructure for interoperable metabolite spectral data and metadata management" (in en). Metabolomics 18 (6): 40. doi:10.1007/s11306-022-01899-3. ISSN 1573-3890. PMC PMC9197906. PMID 35699774. https://link.springer.com/10.1007/s11306-022-01899-3.
- ↑ Berthold, Michael R.; Cebron, Nicolas; Dill, Fabian; Gabriel, Thomas R.; Kötter, Tobias; Meinl, Thorsten; Ohl, Peter; Sieb, Christoph et al.. (2008), Preisach, Christine; Burkhardt, Hans; Schmidt-Thieme, Lars et al.., eds., "KNIME: The Konstanz Information Miner", Data Analysis, Machine Learning and Applications (Berlin, Heidelberg: Springer Berlin Heidelberg): 319–326, doi:10.1007/978-3-540-78246-9_38, ISBN 978-3-540-78239-1, http://link.springer.com/10.1007/978-3-540-78246-9_38. Retrieved 2023-03-13
- ↑ "TeraData". TeraData. 2018. https://www.teradata.com/.
- ↑ "CLC Genomics Workbench". QIAGEN. 2018. Archived from the original on 02 May 2018. https://web.archive.org/web/20180502163500/https://www.qiagenbioinformatics.com/products/clc-genomics-workbench/.
- ↑ Sharma, Vagisha; Eckels, Josh; Taylor, Greg K.; Shulman, Nicholas J.; Stergachis, Andrew B.; Joyner, Shannon A.; Yan, Ping; Whiteaker, Jeffrey R. et al. (5 September 2014). "Panorama: A Targeted Proteomics Knowledge Base" (in en). Journal of Proteome Research 13 (9): 4205–4210. doi:10.1021/pr5006636. ISSN 1535-3893. PMC PMC4156235. PMID 25102069. https://pubs.acs.org/doi/10.1021/pr5006636.
- ↑ "The Seven Bridges Platform". Seven Bridges Genomics. 2018. https://www.sevenbridges.com/platform/.
- ↑ 39.0 39.1 Aleksiev, Tyanko; Barkow-Oesterreicher, Simon; Kunszt, Peter; Maffioletti, Sergio; Murri, Riccardo; Panse, Christian (2013), Kunkel, Julian Martin; Ludwig, Thomas; Meuer, Hans Werner, eds., "VM-MAD: A Cloud/Cluster Software for Service-Oriented Academic Environments" (in en), Supercomputing (Berlin, Heidelberg: Springer Berlin Heidelberg) 7905: 447–461, doi:10.1007/978-3-642-38750-0_34, ISBN 978-3-642-38749-4, https://link.springer.com/10.1007/978-3-642-38750-0_34. Retrieved 2023-03-13
- ↑ Eng, Jimmy K.; Jahan, Tahmina A.; Hoopmann, Michael R. (1 January 2013). "Comet: An open-source MS/MS sequence database search tool" (in en). PROTEOMICS 13 (1): 22–24. doi:10.1002/pmic.201200439. https://onlinelibrary.wiley.com/doi/10.1002/pmic.201200439.
- ↑ "HUPO Proteomics Standards Initiative". Human Proteome Organisation. 2017. https://www.psidev.info/.
- ↑ CRAN Team, Lang, D.T.; Kalibera, T. (3 May 2017). "XML: Tools for Parsing and Generating XML Within R and S-Plus (3.98-1.7)". CRAN. Institute for Statistics and Mathematics of WU. https://cran.r-project.org/package=XML.
- ↑ 43.0 43.1 "fgcz / bfabricShiny (0.11.9)". GitHub. 29 July 2022. https://github.com/fgcz/bfabricShiny.
- ↑ Bilan, Vera; Leutert, Mario; Nanni, Paolo; Panse, Christian; Hottiger, Michael O. (7 February 2017). "Combining Higher-Energy Collision Dissociation and Electron-Transfer/Higher-Energy Collision Dissociation Fragmentation in a Product-Dependent Manner Confidently Assigns Proteomewide ADP-Ribose Acceptor Sites" (in en). Analytical Chemistry 89 (3): 1523–1530. doi:10.1021/acs.analchem.6b03365. ISSN 0003-2700. https://pubs.acs.org/doi/10.1021/acs.analchem.6b03365.
- ↑ 45.0 45.1 45.2 45.3 Trachsel, Christian; Panse, Christian; Kockmann, Tobias; Wolski, Witold E.; Grossmann, Jonas; Schlapbach, Ralph (3 August 2018). "rawDiag: An R Package Supporting Rational LC–MS Method Optimization for Bottom-up Proteomics" (in en). Journal of Proteome Research 17 (8): 2908–2914. doi:10.1021/acs.jproteome.8b00173. ISSN 1535-3893. https://pubs.acs.org/doi/10.1021/acs.jproteome.8b00173.
- ↑ Yoghourdjian, Vahan; Dwyer, Tim; Klein, Karsten; Marriott, Kim; Wybrow, Michael (1 December 2018). "Graph Thumbnails: Identifying and Comparing Multiple Graphs at a Glance". IEEE Transactions on Visualization and Computer Graphics 24 (12): 3081–3095. doi:10.1109/TVCG.2018.2790961. ISSN 1077-2626. https://ieeexplore.ieee.org/document/8249874/.
- ↑ Wickham, Hadley (2009) (in en). ggplot2: Elegant Graphics for Data Analysis. New York, NY: Springer New York. doi:10.1007/978-0-387-98141-3. ISBN 978-0-387-98140-6. https://link.springer.com/10.1007/978-0-387-98141-3.
- ↑ Huber, Wolfgang; Carey, Vincent J; Gentleman, Robert; Anders, Simon; Carlson, Marc; Carvalho, Benilton S; Bravo, Hector Corrada; Davis, Sean et al. (1 February 2015). "Orchestrating high-throughput genomic analysis with Bioconductor" (in en). Nature Methods 12 (2): 115–121. doi:10.1038/nmeth.3252. ISSN 1548-7091. PMC PMC4509590. PMID 25633503. http://www.nature.com/articles/nmeth.3252.
- ↑ Morgan, M.; Shepherd, L., "ExperimentHub: Client to access ExperimentHub resources (2.6.0)", {{{website{{{}}}}}} (BioConductor), doi:10.18129/b9.bioc.experimenthub, https://bioconductor.org/packages/release/bioc/html/ExperimentHub.html
- ↑ 50.0 50.1 Egloff, Pascal; Zimmermann, Iwan; Arnold, Fabian M.; Hutter, Cedric A. J.; Morger, Damien; Opitz, Lennart; Poveda, Lucy; Keserue, Hans-Anton et al. (1 May 2019). "Engineered peptide barcodes for in-depth analyses of binding protein libraries". Nature Methods 16 (5): 421–428. doi:10.1038/s41592-019-0389-8. ISSN 1548-7105. PMC 7116144. PMID 31011184. https://pubmed.ncbi.nlm.nih.gov/31011184.
- ↑ Wolski, Witold E.; Nanni, Paolo; Grossmann, Jonas; d’Errico, Maria; Schlapbach, Ralph; Panse, Christian (9 June 2022) (in en). prolfqua: A Comprehensive R-package for Proteomics Differential Expression Analysis. doi:10.1101/2022.06.07.494524. http://biorxiv.org/lookup/doi/10.1101/2022.06.07.494524.
- ↑ Wilkinson, Mark D.; Dumontier, Michel; Aalbersberg, I. Jsbrand Jan; Appleton, Gabrielle; Axton, Myles; Baak, Arie; Blomberg, Niklas; Boiten, Jan-Willem et al. (15 March 2016). "The FAIR Guiding Principles for scientific data management and stewardship". Scientific Data 3: 160018. doi:10.1038/sdata.2016.18. ISSN 2052-4463. PMC 4792175. PMID 26978244. https://pubmed.ncbi.nlm.nih.gov/26978244.
- ↑ Kockmann, Tobias; Panse, Christian (2 April 2021). "The rawrr R Package: Direct Access to Orbitrap Data and Beyond" (in en). Journal of Proteome Research 20 (4): 2028–2034. doi:10.1021/acs.jproteome.0c00866. ISSN 1535-3893. https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00866.
- ↑ Panse, C.; Grossmann, J.; Barkow-Oesterreicher, S. (4 April 2022). "protViz: Visualizing and Analyzing Mass Spectrometry Related Data in Proteomics (0.7.3)". CRAN. Institute for Statistics and Mathematics of WU. https://cran.r-project.org/package=protViz.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation, grammar, and punctuation. In some cases important information was missing from the references, and that information was added. The authors don't define "core facility" in the original text; a definition and citation is provided for this version. The original Table 1 was turned into a standardized section for abbreviations, which is common to this wiki's format; subsequent tables were incremented upward one for this version.