Journal:Laboratory automation, informatics, and artificial intelligence: Current and future perspectives in clinical microbiology
| Full article title | Laboratory automation, informatics, and artificial intelligence: Current and future perspectives in clinical microbiology |
|---|---|
| Journal | Frontiers in Cellular and Infection Microbiology |
| Author(s) | Mencacci, Antonella; De Socio, Guiseppe V.; Pirelli, Eleonora; Bondi, Paola; Cenci, Elio |
| Author affiliation(s) | University of Perugia, Perugia General Hospital |
| Primary contact | Email: antonella at mencacci at unipg dot it |
| Year published | 2023 |
| Volume and issue | 13 |
| Article # | 1188684 |
| DOI | 10.3389/fcimb.2023.1188684 |
| ISSN | 2235-2988 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.frontiersin.org/articles/10.3389/fcimb.2023.1188684/full |
| Download | https://www.frontiersin.org/articles/10.3389/fcimb.2023.1188684/pdf (PDF) |
|
|
This article should be considered a work in progress and incomplete. Consider this article incomplete until this notice is removed. |
Abstract
Clinical diagnostic laboratories produce one product—information—and for this to be valuable, the information must be clinically relevant, accurate, and timely. Although diagnostic information can clearly improve patient outcomes and decrease healthcare costs, technological challenges and laboratory workflow practices affect the timeliness and clinical value of diagnostics. This article will examine how prioritizing laboratory practices in a patient-oriented approach can be used to optimize technology advances for improved patient care.
Keywords: laboratory automation, artificial intelligence, informatics, laboratory workflow, Kiestra, WASPLab
Introduction
Patterns of infectious diseases have changed dramatically: patients are frequently immunocompromised and often have complicating comorbidities; infections with multi-drug-resistant organisms (MDRO) are a global problem; and new antibiotics are available, but it is mandatory to preserve their efficacy. It is estimated that at least 700,000 people die worldwide every year with infections caused by MDRO, and it is predicted that by 2050, 10 million deaths might occur due to these organisms. [O’Neill, 2016] Administration of rapid, broad-spectrum empiric therapy is essential to improve patient outcome [Levy et al., 2018], but this is often inappropriate. [Kumar et al., 2009; Zilberberg et al., 2017] For example, meta-analysis assessing the impact of antibiotic therapy on Gram-negative sepsis showed that inappropriate therapy was associated with 3.3-fold increased risk of mortality, longer hospitalization, and higher costs. [Raman et al., 2015] Thus, rapid, accurate diagnostics are critical for the selection of the most appropriate therapy.
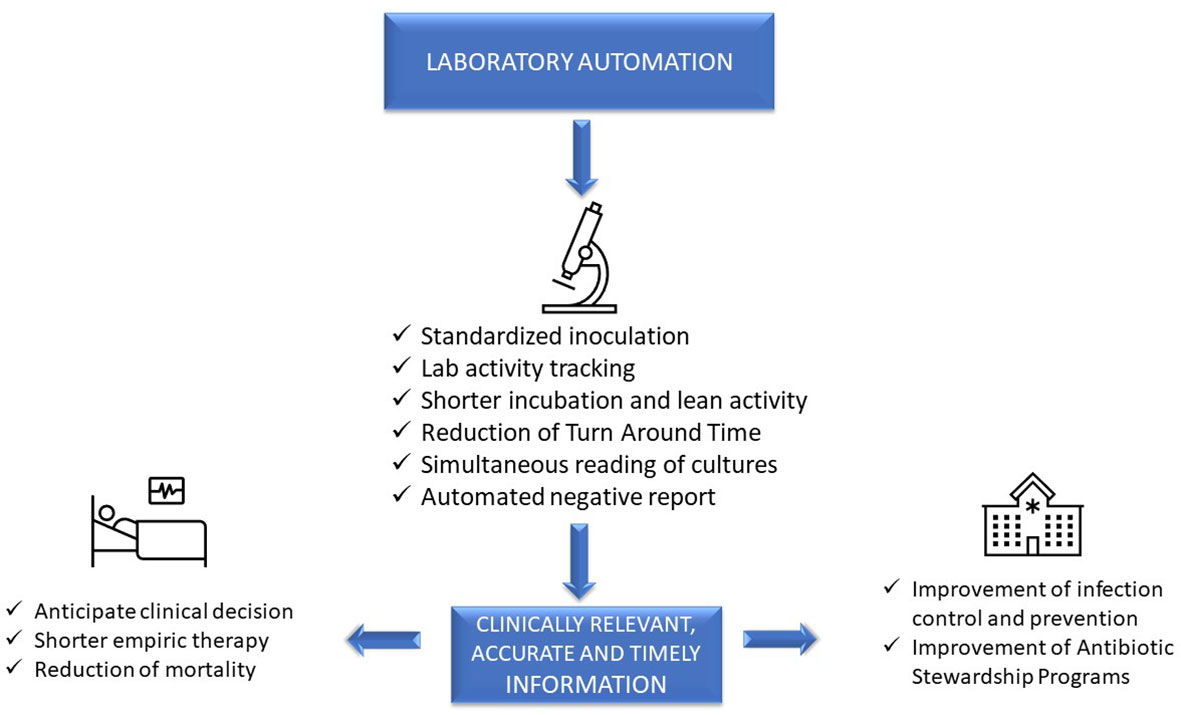
Advanced, sophisticated technologies such as mass spectrometry and molecular diagnostics are rapidly changing our ability to diagnose infections [Trotter et al., 2019], although they should be viewed as complementary to traditional growth-based diagnostics. Laboratory automation and intelligent applications of of informatics also have a transformative impact of microbiology diagnostics. These tools have the potential to accelerate clinical decision-making and positively impact the management of infections, improve patient outcome, and facilitate diagnostic and antimicrobial stewardship (AS) programs. [Messacar et al., 2017] However, it is a challenge for clinical microbiologists to implement these technologies because it requires changing well-established workflow practices. This paper will focus on the impact of automation and informatics combined with workflow changes on laboratory, patient, and hospital management (Figure 1).
|
Impact on laboratory management
References
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. In some cases important information was missing from the references, and that information was added. The original article lists references alphabetically; they are listed by order of appearance for this version, by design.